```{r}
#| code-fold: false
library(ggplot2)
ggplot(data.frame(x=rnorm(100), y=rnorm(100)), aes(x,y)) +
geom_point() +
theme_minimal()
```
An example
Here I want to cite Golder et al. (2022). Maybe I want to cite them in brackets (Golder et al. 2022) or (see Golder et al. 2022, 12). The references are stored in the references.bib file and follow the BibTeX format. You can find BibTeX entries for most papers on google scholar. Below the search entry is a “Cite” button which opens a pop-up that has a “BibTeX” link (e.g., here).
Links to websites can be added to your document with [LINK TEXT](https://example.com).
R code goes into code chunks. Chunk options are set with a special comment #|. Here we added echo: fenced and code-fold: false to show the code but you typically want to remove those from the options:
```{r}
#| code-fold: false
library(ggplot2)
ggplot(data.frame(x=rnorm(100), y=rnorm(100)), aes(x,y)) +
geom_point() +
theme_minimal()
```
Special comments are useful to control output size and captions for images and prevent R from printing warnings (warning: false):
```{r}
#| code-fold: false
#| fig-cap: "Very cool correlation"
#| fig-width: 4
#| fig-height: 4
#| warning: false
library(palmerpenguins)
ggplot(penguins, aes(x = body_mass_g, y = bill_length_mm)) +
geom_point() +
geom_smooth(method="lm") +
theme_minimal()
```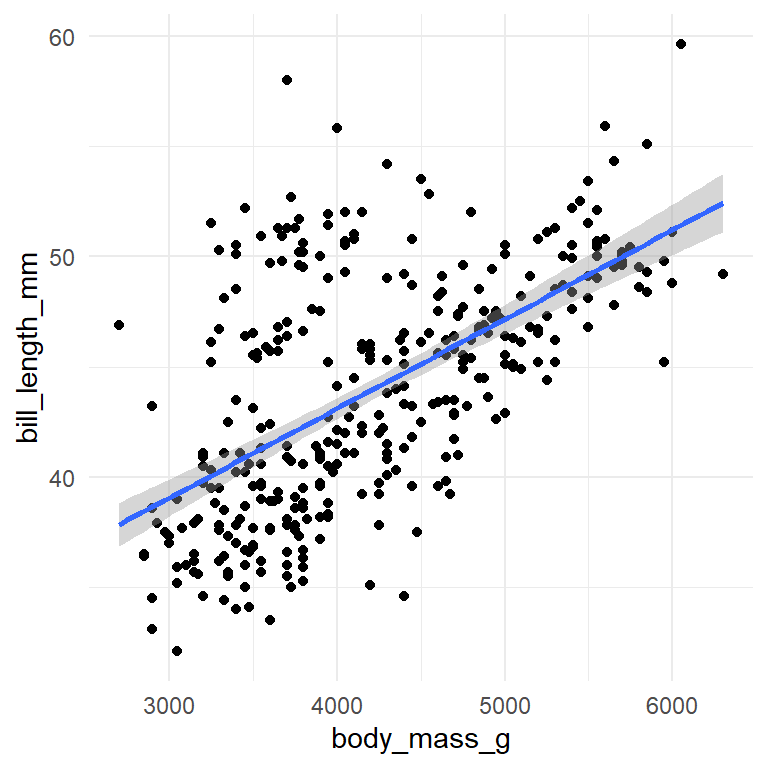
Here is the data
You can use R code inline e.g., to print summary statistics automatically. You could say the average body mass of a penguin in the Palmer Penguins dataset is 4201.8g (`round(mean(penguins$body_mass_g, na.rm=TRUE), digits=1)`).
See the quarto documentation for more info.
library(dplyr)
library(gt)
penguins |>
group_by(island, species) |>
select(-year) |>
summarize_if(is.numeric, \(v)mean(v, na.rm=TRUE)) |>
gt(rowname_col = "species") |>
tab_header(
title = "Penguins",
subtitle = "Average by Island and Species"
) |>
fmt_number(
columns = bill_length_mm:flipper_length_mm,
pattern = "{x}mm",
decimals = 1
) |>
fmt_number(
columns = body_mass_g,
suffixing = "KG",
decimals = 1
) |>
tab_spanner(
label = "Bill",
columns = bill_length_mm:bill_depth_mm
) |>
cols_label(
bill_length_mm = "length",
bill_depth_mm = "depth",
flipper_length_mm = "flipper length",
body_mass_g = "body mass"
)| Penguins | ||||
| Average by Island and Species | ||||
| Bill | flipper length | body mass | ||
|---|---|---|---|---|
| length | depth | |||
| Biscoe | ||||
| Adelie | 39.0mm | 18.4mm | 188.8mm | 3.7KG |
| Gentoo | 47.5mm | 15.0mm | 217.2mm | 5.1KG |
| Dream | ||||
| Adelie | 38.5mm | 18.3mm | 189.7mm | 3.7KG |
| Chinstrap | 48.8mm | 18.4mm | 195.8mm | 3.7KG |
| Torgersen | ||||
| Adelie | 39.0mm | 18.4mm | 191.2mm | 3.7KG |
library(modelsummary)
library(fixest)
## species and year fixed effects with SE clustered by species
## Note that this is JUST for presentation purposes
## and not a meaningful model!!!
mod1 <- feols(
body_mass_g ~ flipper_length_mm + bill_length_mm | species + year,
data = penguins
)
mod2 <- feols(
body_mass_g ~ flipper_length_mm + bill_length_mm + bill_depth_mm | species + year,
data = penguins
)
mod3 <- feols(
body_mass_g ~ flipper_length_mm + bill_length_mm + bill_depth_mm | species,
cluster = ~species,
data = penguins
)
mod4 <- lm(
body_mass_g ~ flipper_length_mm + bill_length_mm + bill_depth_mm + species,
data = penguins
)
modelsummary(
list(`no depth` = mod1,
`with depth` = mod2,
`species FE` = mod3,
`species dummy` = mod4),
coef_map = c(
"flipper_length_mm" = "flipper length",
"bill_length_mm" = "bill length",
"bill_depth_mm" = "bill depth",
"speciesChinstrap" = "Chinstrap vs. Adelie",
"speciesGentoo" = "Gentoo vs. Adelie"
),
gof_map = c(
"nobs",
"r.squared",
"r2.within",
"FE: species",
"FE: year",
"vcov.type"),
vcov = ~species,
title = "Modelling Penguin Body Mass",
notes = "OLS with species dummy results in the same parameters as species FE but is less efficient",
output = "gt"
) |>
tab_spanner(columns = 2:4, label = "Fixed Effects") |>
tab_spanner(columns = 5, label = "OLS")| Fixed Effects | OLS | |||
|---|---|---|---|---|
| no depth | with depth | species FE | species dummy | |
| flipper length | 30.892 | 22.500 | 20.241 | 20.241 |
| (3.999) | (3.853) | (2.690) | (2.698) | |
| bill length | 60.053 | 42.298 | 41.468 | 41.468 |
| (13.244) | (12.217) | (11.604) | (11.638) | |
| bill depth | 131.265 | 140.328 | 140.328 | |
| (16.878) | (16.141) | (16.189) | ||
| Chinstrap vs. Adelie | -513.247 | |||
| (108.516) | ||||
| Gentoo vs. Adelie | 934.887 | |||
| (24.742) | ||||
| Num.Obs. | 342 | 342 | 342 | 342 |
| R2 | 0.829 | 0.849 | 0.847 | 0.847 |
| R2 Within | 0.482 | 0.544 | 0.537 | |
| Std.Errors | by: species | by: species | by: species | by: species |
| FE: species | X | X | X | |
| FE: year | X | X | ||
| OLS with species dummy results in the same parameters as species FE but is less efficient | ||||