Difference in Differences Workshop
UNSW Data Science Hub
Preliminaries
This tutorial uses the R statistical software (R Core Team 2022) and assumes you have a recent version with the standard packages loaded (this is the case in a fresh installation without modifications).
Code
library(ggplot2)
library(psych)
library(data.table)
library(tidyverse)
options(scipen = 999)
# panel data models
#-------------------------------------------------------------------
# load data
music_data <- fread("https://raw.githubusercontent.com/WU-RDS/RMA2022/main/data/music_data.csv")
# convert to factor
music_data$song_id <- as.factor(music_data$song_id)
music_data$genre <- as.factor(music_data$genre)
music_dataCode
# example plot to visualize the data structure
ggplot(music_data, aes(x = week, y = streams / 1000000, group = song_id, fill = song_id, color = song_id)) +
geom_area(position = "stack", alpha = 0.65) +
labs(x = "Week", y = "Total streams (in million)", title = "Weekly number of streams by song") +
theme_bw() +
theme(plot.title = element_text(hjust = 0.5, color = "#666666"), legend.position = "none")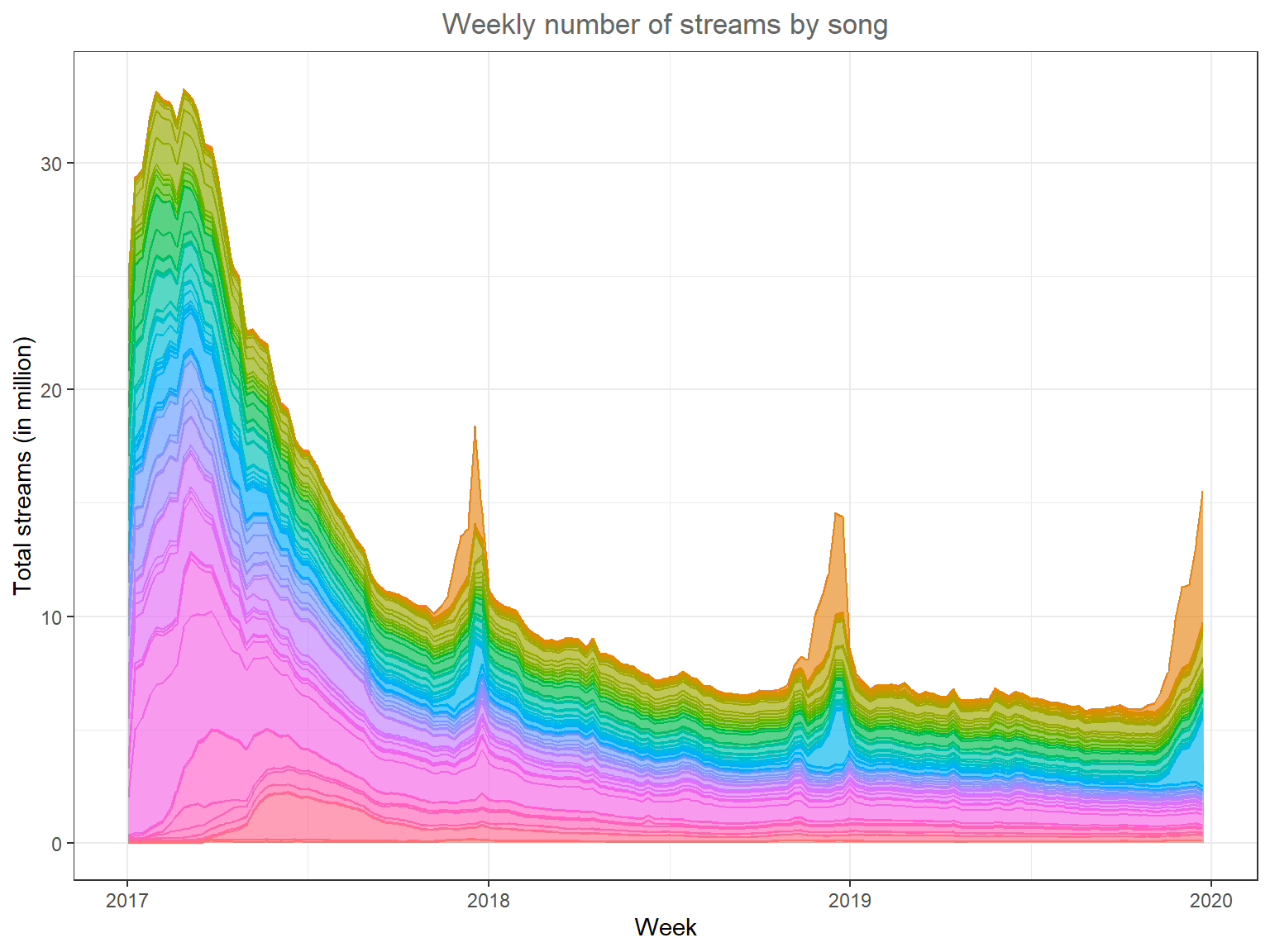
Baseline model
Estimation of different specifications of a linear model using OLS (without fixed effects). The lm command for OLS estimation is available in your R session by default.
Code
# estimate the baseline model
fe_m0 <- lm(log(streams) ~ log(radio + 1) + log(adspend + 1),
data = music_data
)
# ... + control for song age
fe_m1 <- lm(log(streams) ~ log(radio + 1) + log(adspend + 1) + log(weeks_since_release + 1),
data = music_data
)
# ... + playlist follower variable
fe_m2 <- lm(log(streams) ~ log(radio + 1) + log(adspend + 1) + log(weeks_since_release + 1) + log(playlist_follower),
data = music_data
)
# ... + song fixed effects
fe_m3 <- lm(
log(streams) ~ log(radio + 1) + log(adspend + 1) + log(playlist_follower) + log(weeks_since_release + 1) +
as.factor(song_id),
data = music_data
)
library(stargazer)
stargazer(fe_m0, fe_m1, fe_m2, fe_m3, type = "html")| Dependent variable: | ||||
| log(streams) | ||||
| (1) | (2) | (3) | (4) | |
| log(radio + 1) | 0.542*** | 0.503*** | 0.280*** | 0.107*** |
| (0.012) | (0.013) | (0.010) | (0.004) | |
| log(adspend + 1) | 0.033*** | 0.035*** | 0.056*** | 0.022*** |
| (0.011) | (0.011) | (0.008) | (0.003) | |
| log(weeks_since_release + 1) | -0.143*** | 0.074*** | -0.400*** | |
| (0.014) | (0.010) | (0.008) | ||
| as.factor(song_id)2 | 0.918*** | |||
| (0.051) | ||||
| as.factor(song_id)3 | 0.026 | |||
| (0.051) | ||||
| as.factor(song_id)4 | 1.257*** | |||
| (0.050) | ||||
| as.factor(song_id)5 | -0.336*** | |||
| (0.051) | ||||
| as.factor(song_id)6 | 0.648*** | |||
| (0.051) | ||||
| as.factor(song_id)7 | 2.641*** | |||
| (0.061) | ||||
| as.factor(song_id)8 | 3.802*** | |||
| (0.052) | ||||
| as.factor(song_id)9 | 3.960*** | |||
| (0.051) | ||||
| as.factor(song_id)10 | 3.812*** | |||
| (0.054) | ||||
| as.factor(song_id)11 | 2.681*** | |||
| (0.055) | ||||
| as.factor(song_id)12 | 4.046*** | |||
| (0.052) | ||||
| as.factor(song_id)13 | 2.976*** | |||
| (0.050) | ||||
| as.factor(song_id)14 | 0.727*** | |||
| (0.053) | ||||
| as.factor(song_id)15 | 4.031*** | |||
| (0.050) | ||||
| as.factor(song_id)16 | 4.032*** | |||
| (0.054) | ||||
| as.factor(song_id)17 | 4.422*** | |||
| (0.052) | ||||
| as.factor(song_id)18 | 0.915*** | |||
| (0.052) | ||||
| as.factor(song_id)19 | 2.971*** | |||
| (0.050) | ||||
| as.factor(song_id)20 | 2.334*** | |||
| (0.061) | ||||
| as.factor(song_id)21 | 4.930*** | |||
| (0.053) | ||||
| as.factor(song_id)22 | 3.401*** | |||
| (0.057) | ||||
| as.factor(song_id)23 | 3.040*** | |||
| (0.060) | ||||
| as.factor(song_id)24 | 2.695*** | |||
| (0.060) | ||||
| as.factor(song_id)25 | 1.229*** | |||
| (0.050) | ||||
| as.factor(song_id)26 | 4.313*** | |||
| (0.052) | ||||
| as.factor(song_id)27 | 3.514*** | |||
| (0.055) | ||||
| as.factor(song_id)28 | 1.026*** | |||
| (0.055) | ||||
| as.factor(song_id)29 | 4.440*** | |||
| (0.051) | ||||
| as.factor(song_id)30 | 2.444*** | |||
| (0.062) | ||||
| as.factor(song_id)31 | 0.615*** | |||
| (0.055) | ||||
| as.factor(song_id)32 | 1.218*** | |||
| (0.054) | ||||
| as.factor(song_id)33 | -0.246*** | |||
| (0.052) | ||||
| as.factor(song_id)34 | 3.587*** | |||
| (0.055) | ||||
| as.factor(song_id)35 | 4.965*** | |||
| (0.054) | ||||
| as.factor(song_id)36 | 1.904*** | |||
| (0.053) | ||||
| as.factor(song_id)37 | 1.255*** | |||
| (0.054) | ||||
| as.factor(song_id)38 | 2.409*** | |||
| (0.062) | ||||
| as.factor(song_id)39 | 0.375*** | |||
| (0.053) | ||||
| as.factor(song_id)40 | 2.616*** | |||
| (0.050) | ||||
| as.factor(song_id)41 | 0.803*** | |||
| (0.053) | ||||
| as.factor(song_id)42 | 1.569*** | |||
| (0.054) | ||||
| as.factor(song_id)43 | 2.041*** | |||
| (0.053) | ||||
| as.factor(song_id)44 | 1.517*** | |||
| (0.051) | ||||
| as.factor(song_id)45 | 2.792*** | |||
| (0.064) | ||||
| as.factor(song_id)46 | 3.707*** | |||
| (0.051) | ||||
| as.factor(song_id)47 | 2.526*** | |||
| (0.061) | ||||
| as.factor(song_id)48 | 2.781*** | |||
| (0.051) | ||||
| as.factor(song_id)49 | 1.731*** | |||
| (0.054) | ||||
| as.factor(song_id)50 | 1.655*** | |||
| (0.058) | ||||
| as.factor(song_id)51 | 2.293*** | |||
| (0.061) | ||||
| as.factor(song_id)52 | 0.406*** | |||
| (0.052) | ||||
| as.factor(song_id)53 | 2.113*** | |||
| (0.060) | ||||
| as.factor(song_id)54 | 3.615*** | |||
| (0.051) | ||||
| as.factor(song_id)55 | 3.393*** | |||
| (0.052) | ||||
| as.factor(song_id)56 | 1.346*** | |||
| (0.056) | ||||
| as.factor(song_id)57 | 2.891*** | |||
| (0.056) | ||||
| as.factor(song_id)58 | 2.277*** | |||
| (0.052) | ||||
| as.factor(song_id)59 | 1.621*** | |||
| (0.056) | ||||
| as.factor(song_id)60 | 3.518*** | |||
| (0.059) | ||||
| as.factor(song_id)61 | 0.906*** | |||
| (0.051) | ||||
| as.factor(song_id)62 | 1.741*** | |||
| (0.058) | ||||
| as.factor(song_id)63 | 2.100*** | |||
| (0.052) | ||||
| as.factor(song_id)64 | 2.151*** | |||
| (0.060) | ||||
| as.factor(song_id)65 | 0.438*** | |||
| (0.051) | ||||
| as.factor(song_id)66 | 3.008*** | |||
| (0.059) | ||||
| as.factor(song_id)67 | 2.116*** | |||
| (0.063) | ||||
| as.factor(song_id)68 | 2.413*** | |||
| (0.061) | ||||
| as.factor(song_id)69 | 2.247*** | |||
| (0.053) | ||||
| as.factor(song_id)70 | 2.829*** | |||
| (0.059) | ||||
| as.factor(song_id)71 | 2.523*** | |||
| (0.055) | ||||
| as.factor(song_id)72 | -0.802*** | |||
| (0.051) | ||||
| as.factor(song_id)73 | 2.950*** | |||
| (0.059) | ||||
| as.factor(song_id)74 | 1.618*** | |||
| (0.051) | ||||
| as.factor(song_id)75 | 3.039*** | |||
| (0.055) | ||||
| as.factor(song_id)76 | 4.060*** | |||
| (0.051) | ||||
| as.factor(song_id)77 | 4.201*** | |||
| (0.053) | ||||
| as.factor(song_id)78 | 2.814*** | |||
| (0.062) | ||||
| as.factor(song_id)79 | 3.514*** | |||
| (0.051) | ||||
| as.factor(song_id)80 | 1.954*** | |||
| (0.056) | ||||
| as.factor(song_id)81 | 2.912*** | |||
| (0.062) | ||||
| as.factor(song_id)82 | 3.188*** | |||
| (0.065) | ||||
| as.factor(song_id)83 | 2.051*** | |||
| (0.053) | ||||
| as.factor(song_id)84 | 0.161*** | |||
| (0.051) | ||||
| as.factor(song_id)85 | -0.172*** | |||
| (0.052) | ||||
| as.factor(song_id)86 | 2.882*** | |||
| (0.060) | ||||
| as.factor(song_id)87 | 2.757*** | |||
| (0.051) | ||||
| as.factor(song_id)88 | 3.023*** | |||
| (0.058) | ||||
| as.factor(song_id)89 | 3.063*** | |||
| (0.062) | ||||
| as.factor(song_id)90 | 1.302*** | |||
| (0.053) | ||||
| as.factor(song_id)91 | 2.458*** | |||
| (0.062) | ||||
| as.factor(song_id)92 | 1.426*** | |||
| (0.054) | ||||
| as.factor(song_id)93 | 0.672*** | |||
| (0.051) | ||||
| as.factor(song_id)94 | 2.901*** | |||
| (0.062) | ||||
| as.factor(song_id)95 | 0.710*** | |||
| (0.052) | ||||
| as.factor(song_id)96 | 0.944*** | |||
| (0.058) | ||||
| as.factor(song_id)97 | 4.557*** | |||
| (0.052) | ||||
| log(playlist_follower) | 0.536*** | 0.439*** | ||
| (0.005) | (0.006) | |||
| Constant | 10.091*** | 10.800*** | 2.530*** | 3.938*** |
| (0.015) | (0.070) | (0.091) | (0.083) | |
| Observations | 15,132 | 15,132 | 15,132 | 15,132 |
| R2 | 0.130 | 0.136 | 0.524 | 0.945 |
| Adjusted R2 | 0.130 | 0.136 | 0.523 | 0.944 |
| Residual Std. Error | 1.734 (df = 15129) | 1.728 (df = 15128) | 1.283 (df = 15127) | 0.439 (df = 15031) |
| F Statistic | 1,129.772*** (df = 2; 15129) | 794.258*** (df = 3; 15128) | 4,156.097*** (df = 4; 15127) | 2,564.991*** (df = 100; 15031) |
| Note: | p<0.1; p<0.05; p<0.01 | |||
Fixed Effects model
Estimation of a linear model with song and song-time fixed effects using the fixest (Bergé 2018) package. Note that this package also implements the estimator proposed by Sun and Abraham (2020) for staggered adoption (see ?sunab).
Code
# ... same as m3 using the fixest package
library(fixest) # https://lrberge.github.io/fixest/
fe_m4 <- feols(
log(streams) ~ log(radio + 1) + log(adspend + 1) + log(playlist_follower) + log(weeks_since_release + 1) |
song_id,
data = music_data
)
# ... + week fixed effects
fe_m5 <- feols(
log(streams) ~ log(radio + 1) + log(adspend + 1) + log(playlist_follower) + log(weeks_since_release + 1) |
song_id + week,
data = music_data
)
etable(fe_m4, fe_m5, se = "cluster")Code
# extract fixed effects coefficients
fixed_effects <- fixef(fe_m5)
summary(fixed_effects)Fixed_effects coefficients
song_id week
Number of fixed-effects 97 156
Number of references 0 1
Mean 6.37 0.688
Standard-deviation 1.37 0.161
COEFFICIENTS:
song_id: 1 2 3 4 5
4.188 5.041 4.061 5.27 3.66 ... 92 remaining
-----
week: 2017-01-02 2017-01-09 2017-01-16 2017-01-23 2017-01-30
0 0.0499 0.06933 0.171 0.221
... 151 remainingCode
plot(fixed_effects)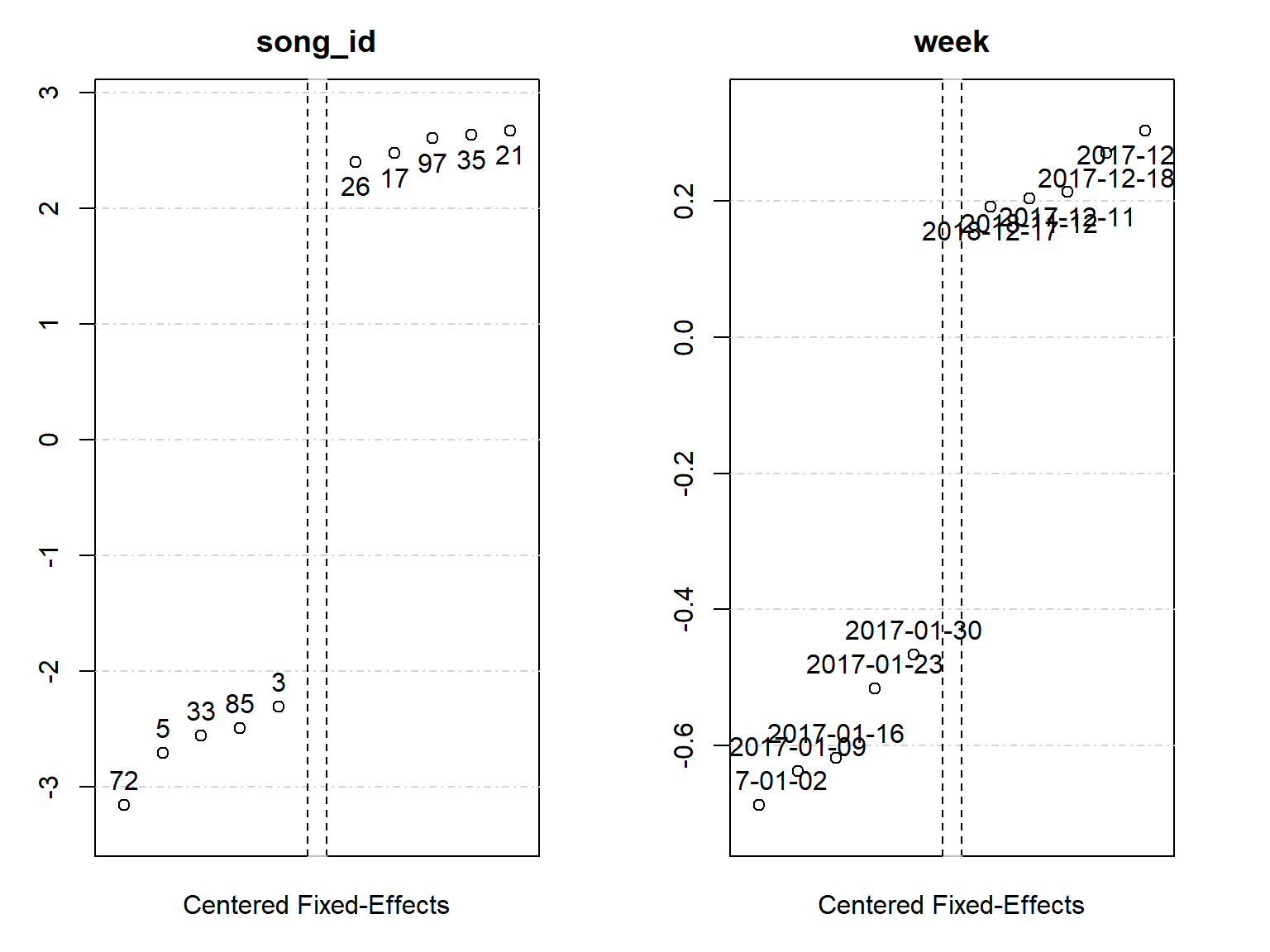
Mixed Effects model
The term Fixed Effects is not used consistently in the literature. Here we view fixed effects as unit level constants (R packages: fixest, lfe, or alpaca). If you find the term “fixed effects” in the context of “mixed effects models” or “random effects models” (in R when the lme4 package is used) it refers to coefficients that are fixed across units.
Estimation of mixed effects model using lme4. This allows explicitly modelling heterogeneity of intercept and slopes across units.
Code
library(lme4) # https://github.com/lme4/lme4
music_data$log_playlist_follower <- log(music_data$playlist_follower)
music_data$log_streams <- log(music_data$streams)
re_m1 <- lmer(log_streams ~ log(radio + 1) + log(adspend + 1) + log_playlist_follower + log(weeks_since_release + 1) + (1 + log_playlist_follower | song_id), data = music_data)
summary(re_m1)Linear mixed model fit by REML ['lmerMod']
Formula:
log_streams ~ log(radio + 1) + log(adspend + 1) + log_playlist_follower +
log(weeks_since_release + 1) + (1 + log_playlist_follower | song_id)
Data: music_data
REML criterion at convergence: 14776.3
Scaled residuals:
Min 1Q Median 3Q Max
-8.4922 -0.3578 -0.0084 0.3620 13.7550
Random effects:
Groups Name Variance Std.Dev. Corr
song_id (Intercept) 26.467 5.1446
log_playlist_follower 0.140 0.3742 -0.95
Residual 0.144 0.3794
Number of obs: 15132, groups: song_id, 97
Fixed effects:
Estimate Std. Error t value
(Intercept) 6.369815 0.537152 11.86
log(radio + 1) 0.084025 0.004081 20.59
log(adspend + 1) 0.026216 0.002496 10.50
log_playlist_follower 0.446280 0.038877 11.48
log(weeks_since_release + 1) -0.478857 0.008095 -59.16
Correlation of Fixed Effects:
(Intr) lg(r+1) lg(d+1) lg_pl_
log(radi+1) 0.051
lg(dspnd+1) 0.011 -0.126
lg_plylst_f -0.952 -0.073 -0.013
lg(wks__+1) -0.076 0.382 0.043 0.000Code
library(sjPlot)
plot_model(re_m1, show.values = TRUE, value.offset = .3)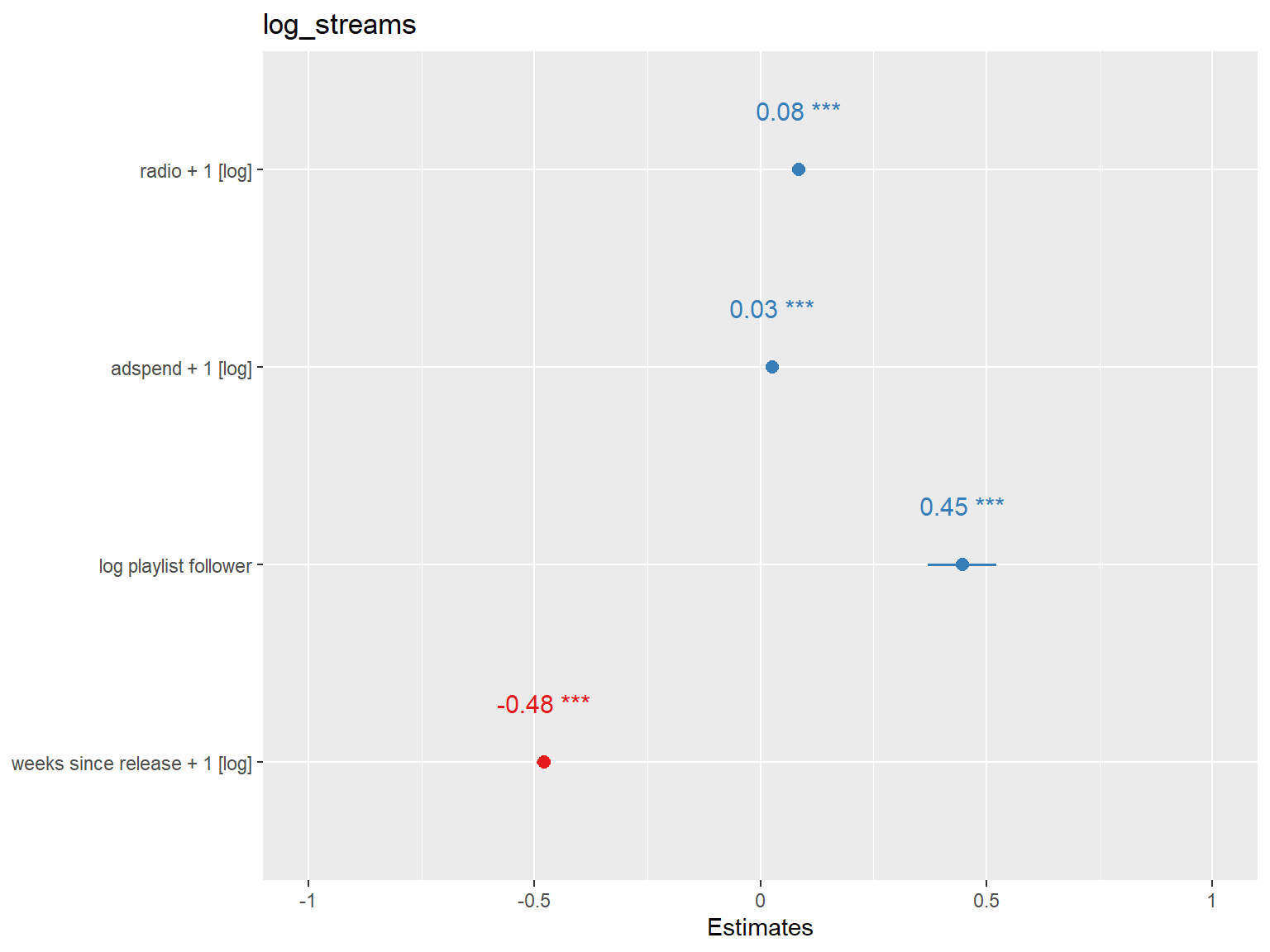
Code
# plot random-slope-intercept
plot_model(re_m1,
type = "pred", terms = c("log_playlist_follower", "song_id"),
pred.type = "re", ci.lvl = NA
) +
scale_colour_manual(values = hcl(0, 100, seq(40, 100, length = 97))) +
theme(legend.position = "bottom", legend.key.size = unit(0.3, "cm")) + guides(colour = guide_legend(nrow = 5))
Difference-in-Differences estimator
“Canonical” Diff-in-Diff
Data preparation using the data.table and dplyr packages.
Code
# load data
did_data <- fread("https://raw.github.com/WU-RDS/RMA2022/main/data/did_data_exp.csv")
# pre-processing
did_data$song_id <- as.character(did_data$song_id)
did_data$treated_fct <- factor(did_data$treated, levels = c(0, 1), labels = c("non-treated", "treated"))
did_data$post_fct <- factor(did_data$post, levels = c(0, 1), labels = c("pre", "post"))
did_data$week <- as.Date(did_data$week)
did_data <- did_data %>%
dplyr::filter(!song_id %in% c("101", "143", "154", "63", "161", "274")) %>%
as.data.frame()
did_dataNumber of songs by treatment status:
Code
library(gt)
did_data %>%
dplyr::group_by(treated) %>%
dplyr::summarise(unique_songs = n_distinct(song_id)) |>
gt()| treated | unique_songs |
|---|---|
| 0 | 231 |
| 1 | 53 |
Visualization of the panel structure using the panelView package.
Code
head(did_data)Code
did_data %>%
dplyr::group_by(treated) %>%
dplyr::summarise(unique_songs = n_distinct(song_id))Code
library(panelView) # https://yiqingxu.org/packages/panelview/
panelview(streams ~ treated_post, data = did_data, index = c("song_id", "week"), pre.post = TRUE, by.timing = TRUE, axis.lab.angle = 90)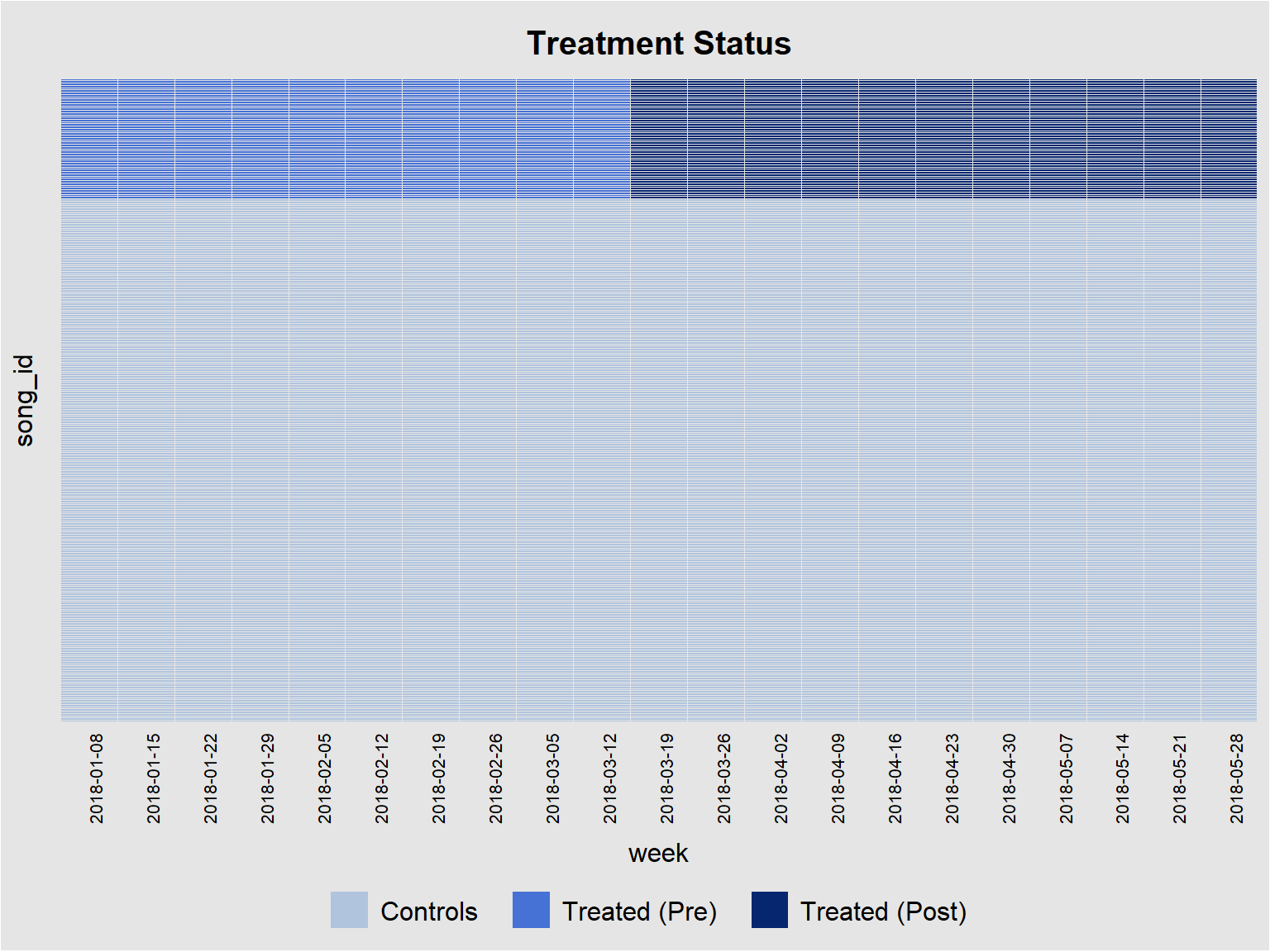
Code
panelview(streams ~ treated_post, data = did_data, index = c("song_id", "week"), type = "outcome", axis.lab.angle = 90)
Code
did_data <- did_data %>%
group_by(song_id) %>%
dplyr::mutate(mean_streams = mean(streams)) %>%
as.data.frame()
panelview(streams ~ treated_post, data = did_data %>% dplyr::filter(mean_streams < 70000), index = c("song_id", "week"), type = "outcome", axis.lab.angle = 90)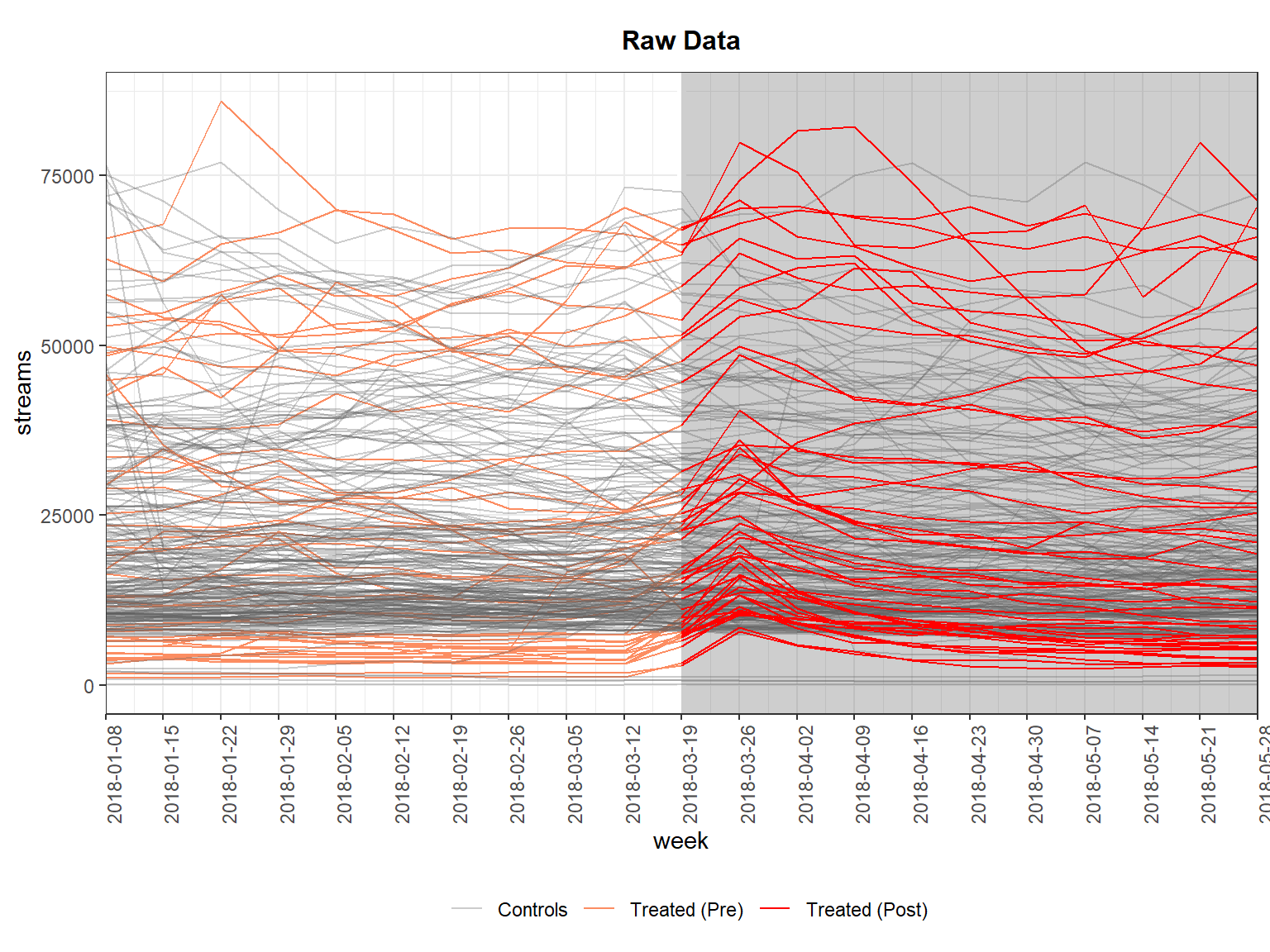
Visualization of the outcome variable by treatment group using the ggplot2 package.
Code
# alternatively, split plot by group
# compute the mean streams per group and week
did_data <- did_data %>%
dplyr::group_by(treated_fct, week) %>%
dplyr::mutate(mean_streams_grp = mean(log(streams))) %>%
as.data.frame()
# set color scheme for songs
cols <- c(rep("gray", length(unique(did_data$song_id))))
# set labels for axis
abbrev_x <- c(
"-10", "", "-8", "",
"-6", "", "-4", "",
"-2", "", "0",
"", "+2", "", "+4",
"", "+6", "", "+8",
"", "+10"
)
# axis titles and names
title_size <- 24
font_size <- 14
line_size <- 1 / 2
# create plot
ggplot(did_data) +
geom_step(aes(x = week, y = log(streams), color = song_id), alpha = 0.75) +
geom_step(aes(x = week, y = mean_streams_grp), color = "black", size = 2, alpha = 0.5) +
# geom_vline(xintercept = as.Date("2018-03-19"),color="black",linetype="dashed") +
labs(
x = "week before/after playlist listing", y = "ln(streams)",
title = "Number of weekly streams per song"
) +
scale_color_manual(values = cols) +
theme_bw() +
scale_x_continuous(breaks = unique(did_data$week), labels = abbrev_x) +
theme(
legend.position = "none",
panel.grid.minor.x = element_blank(),
panel.grid.major.x = element_blank(),
strip.text.x = element_text(size = font_size),
panel.grid.major.y = element_line(
color = "gray75",
size = 0.25,
linetype = 1
),
panel.grid.minor.y = element_line(
color = "gray75",
size = 0.25,
linetype = 1
),
plot.title = element_text(color = "#666666", size = title_size),
axis.title = element_text(size = font_size),
axis.text = element_text(size = font_size),
plot.subtitle = element_text(color = "#666666", size = font_size),
axis.text.x = element_text(size = font_size)
) +
facet_wrap(~treated_fct)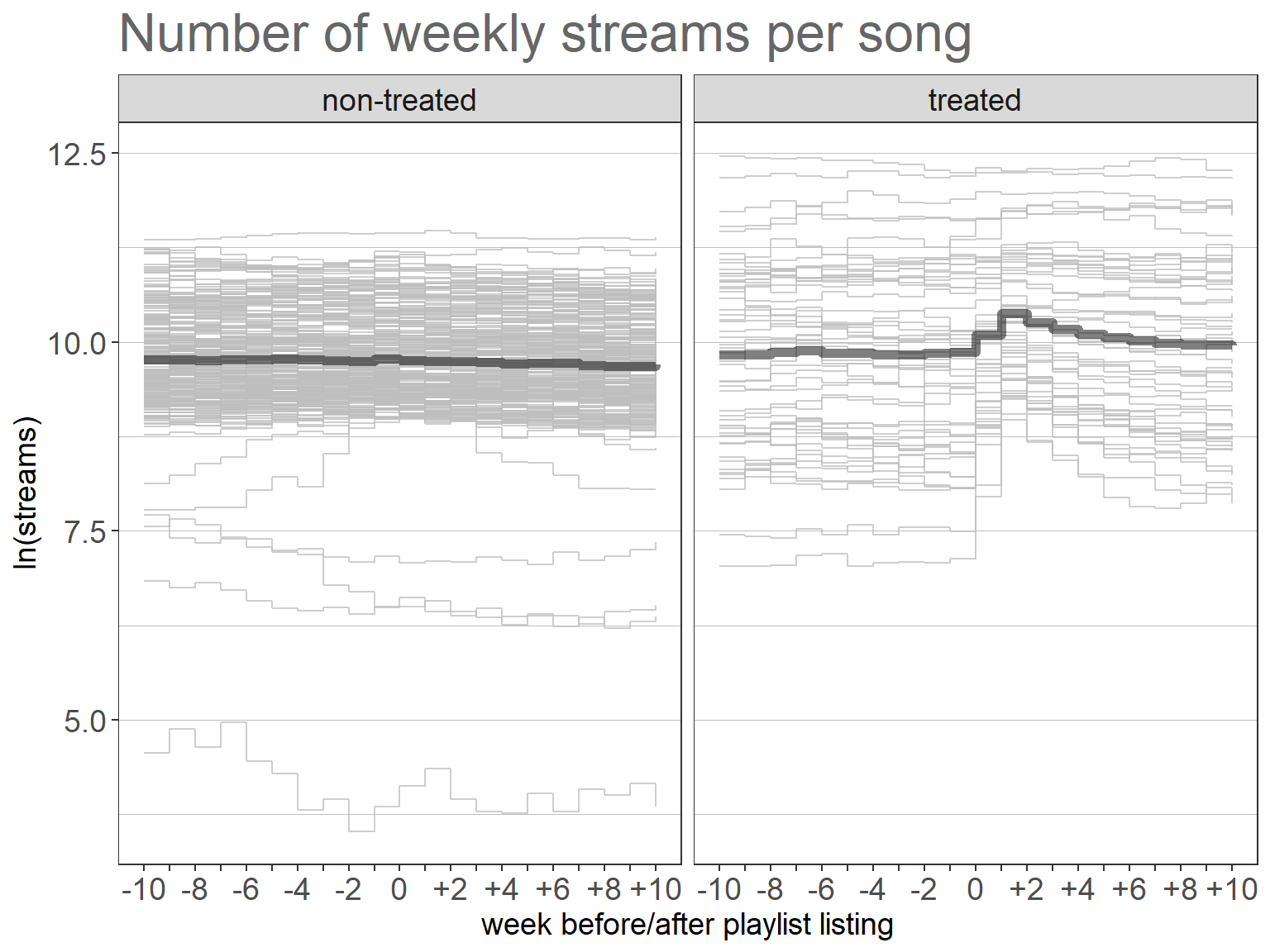
Baseline model
Estimation of the baseline model using dummy-variables for the post-treatment period (\(1\) for all units in the post-treatment period) and the treatment group (always \(1\) for units that are eventually treated) as well as their interaction. The coefficient of the latter is the difference-in-differences estimator of the treatment effect.
Code
# run baseline did model
did_m1 <- lm(log(streams + 1) ~ treated * post,
data = did_data %>% dplyr::filter(week != as.Date("2018-03-19"))
)
stargazer(did_m1, type = "html")| Dependent variable: | |
| log(streams + 1) | |
| treated | 0.086** |
| (0.041) | |
| post | -0.053** |
| (0.025) | |
| treated:post | 0.287*** |
| (0.058) | |
| Constant | 9.767*** |
| (0.018) | |
| Observations | 5,680 |
| R2 | 0.015 |
| Adjusted R2 | 0.014 |
| Residual Std. Error | 0.857 (df = 5676) |
| F Statistic | 28.782*** (df = 3; 5676) |
| Note: | p<0.1; p<0.05; p<0.01 |
Two-Way Fixed Effects model
Alternative estimation of the same difference-in-differences parameter as above using unit-time fixed effects (fixest).
Code
# same model using fixed effects specification
did_m2 <- feols(
log(streams + 1) ~ treated * post |
song_id + week,
cluster = "song_id",
data = did_data %>% dplyr::filter(week != as.Date("2018-03-19"))
)
etable(did_m2, se = "cluster")The did package
One of the most useful packages for the estimation of difference-in-differences with staggered adoption is did (Callaway and Sant’Anna 2021a). However the approach proposed by Callaway and Sant’Anna (2021b) also naturally extends to comparison of the canonical diff-in-diffs specification. First, group-time ATTs are calculated (in their paper group refers to adoption cohort). Second, the ATTs are aggregated using a group-size weighted average. Inference for this aggregate is implemented in the package.
Code
### DID PACKAGE
library(did)
## Treatment happens between period 10 & 11
## G is indicator of when a unit is treated
did_data$period <- as.numeric(factor(as.character(did_data$week),
levels = as.character(sort(unique(did_data$week)))
))
did_data$log_streams <- log(did_data$streams + 1)
did_data$G <- 11 * did_data$treated
did_data$id <- as.numeric(did_data$song_id)
simple_gt_att <- att_gt(
yname = "log_streams",
tname = "period",
idname = "id",
gname = "G",
data = did_data
)
## "simple" gives a weighted average of group-time effects weighted by group (treatment time) size
aggte(simple_gt_att, type = "simple")
Call:
aggte(MP = simple_gt_att, type = "simple")
Reference: Callaway, Brantly and Pedro H.C. Sant'Anna. "Difference-in-Differences with Multiple Time Periods." Journal of Econometrics, Vol. 225, No. 2, pp. 200-230, 2021. <https://doi.org/10.1016/j.jeconom.2020.12.001>, <https://arxiv.org/abs/1803.09015>
ATT Std. Error [ 95% Conf. Int.]
0.2813 0.0401 0.2028 0.3599 *
---
Signif. codes: `*' confidence band does not cover 0
Control Group: Never Treated, Anticipation Periods: 0
Estimation Method: Doubly RobustParallel trends assessment
Period-specific effects
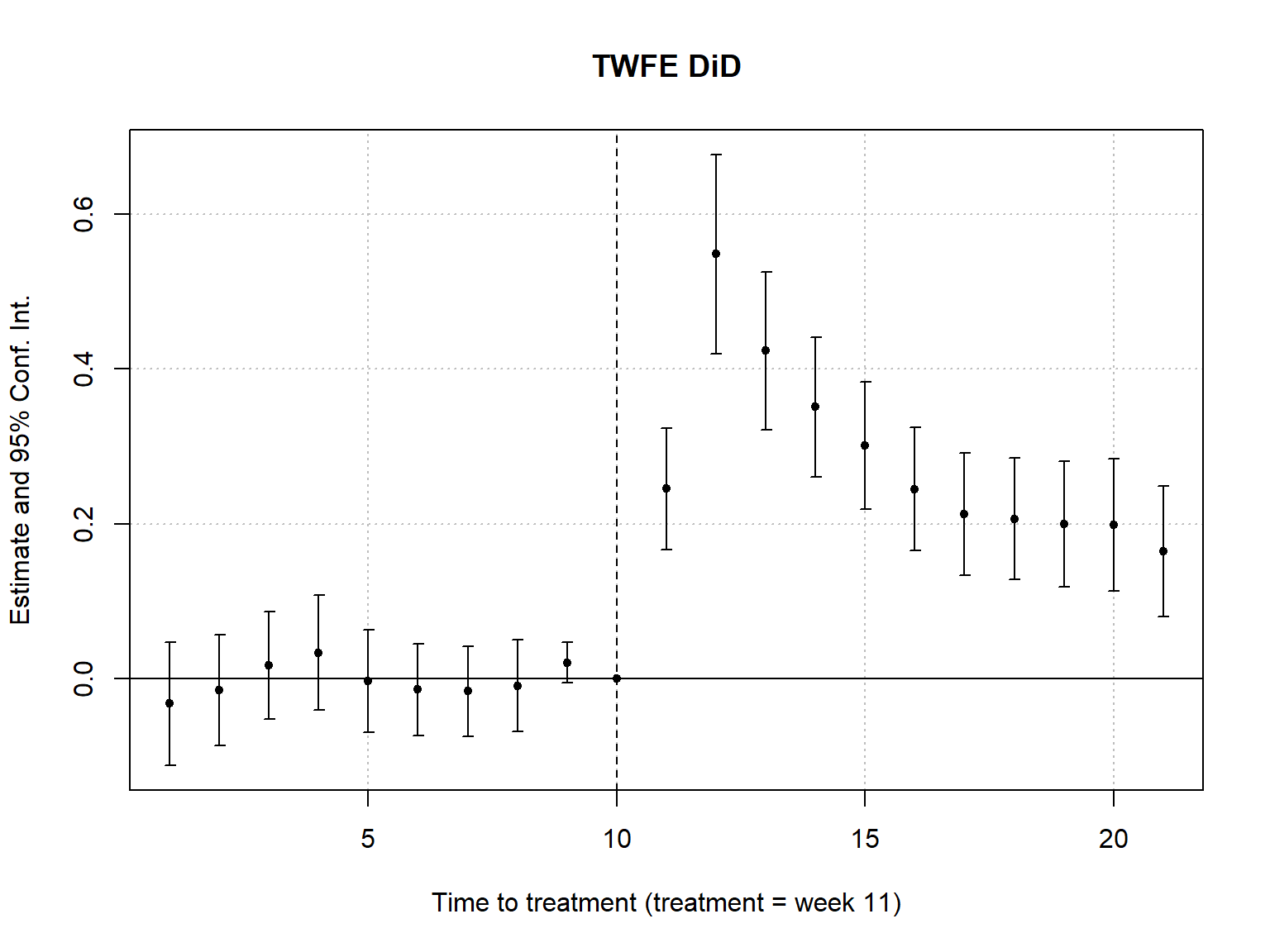
Conduct leads & lags analysis to assess “pre-trends” using fixest. \(H_0\) should not be rejected pre-treatment.
Code
did_data <- did_data %>%
dplyr::group_by(song_id) %>%
dplyr::mutate(period = seq(n())) %>%
as.data.frame()
did_m3 <- fixest::feols(log(streams) ~ i(period, treated, ref = 10) | song_id + period,
cluster = "song_id",
data = did_data
)
etable(did_m3, se = "cluster")Code
fixest::iplot(did_m3,
xlab = "Time to treatment (treatment = week 11)",
main = "TWFE DiD"
)
did provides a Wald test for pre-treatment trends.
Code
## for time < group these are pseudo ATTs -> pre-test for parallel trends
summary(simple_gt_att)
Call:
att_gt(yname = "log_streams", tname = "period", idname = "id",
gname = "G", data = did_data)
Reference: Callaway, Brantly and Pedro H.C. Sant'Anna. "Difference-in-Differences with Multiple Time Periods." Journal of Econometrics, Vol. 225, No. 2, pp. 200-230, 2021. <https://doi.org/10.1016/j.jeconom.2020.12.001>, <https://arxiv.org/abs/1803.09015>
Group-Time Average Treatment Effects:
Group Time ATT(g,t) Std. Error [95% Simult. Conf. Band]
11 2 0.0176 0.0157 -0.0234 0.0586
11 3 0.0316 0.0138 -0.0045 0.0677
11 4 0.0168 0.0161 -0.0251 0.0586
11 5 -0.0369 0.0130 -0.0708 -0.0030 *
11 6 -0.0108 0.0153 -0.0508 0.0292
11 7 -0.0022 0.0105 -0.0296 0.0252
11 8 0.0070 0.0139 -0.0291 0.0432
11 9 0.0296 0.0305 -0.0499 0.1092
11 10 -0.0200 0.0138 -0.0559 0.0159
11 11 0.2449 0.0412 0.1376 0.3523 *
11 12 0.5485 0.0658 0.3769 0.7202 *
11 13 0.4236 0.0535 0.2842 0.5630 *
11 14 0.3508 0.0481 0.2254 0.4762 *
11 15 0.3010 0.0429 0.1892 0.4127 *
11 16 0.2445 0.0408 0.1381 0.3509 *
11 17 0.2123 0.0410 0.1055 0.3191 *
11 18 0.2063 0.0411 0.0990 0.3135 *
11 19 0.1994 0.0427 0.0881 0.3108 *
11 20 0.1988 0.0437 0.0849 0.3127 *
11 21 0.1646 0.0434 0.0513 0.2778 *
---
Signif. codes: `*' confidence band does not cover 0
P-value for pre-test of parallel trends assumption: 0.12298
Control Group: Never Treated, Anticipation Periods: 0
Estimation Method: Doubly RobustCode
ggdid(simple_gt_att)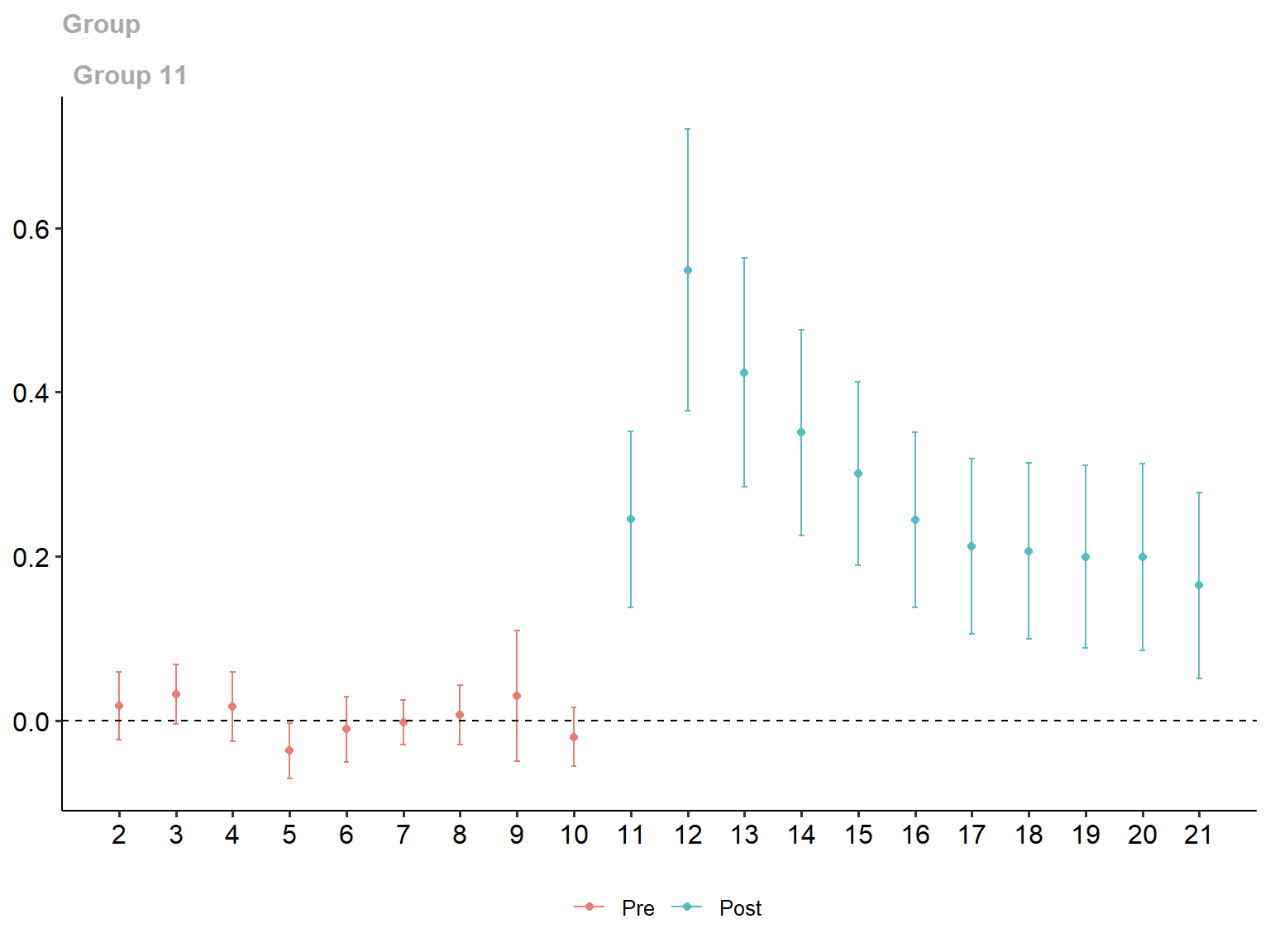
Placebo-test
Code
did_data_placebo <- did_data %>%
dplyr::filter(period < 11) %>%
as.data.frame()
did_data_placebo$post <- ifelse(did_data_placebo$period >= 5, 1, 0)
placebo_model <- feols(
log(streams + 1) ~ treated * post |
song_id + week,
cluster = "song_id",
data = did_data_placebo
)
etable(placebo_model, se = "cluster")Sensitivity analysis
Rambachan and Roth (2023) propose (among other things) to test the robustness of the results to violations of the parallel trends in the post-treatment period relative to the pre-treatment period. We can check statistical significance of causale parameters if we impose that the post-treatment violation of parallel trends is not more than \(\bar M\) longer than the worst pre-treatment violation of parallel trends for different values of \(\bar M\).
Code
##### - honest pre-treatment trend assessment; https://github.com/asheshrambachan/HonestDiD
library(Rglpk)
library(HonestDiD)
betahat <- summary(did_m3)$coefficients # save the coefficients
sigma <- summary(did_m3)$cov.scaled # save the covariance matrix
## significant result is robust to allowing for violations of parallel trends
## up to >twice as big as the max violation in the pre-treatment period.
HonestDiD::createSensitivityResults_relativeMagnitudes(
betahat = betahat, # coefficients
sigma = sigma, # covariance matrix
numPrePeriods = 10, # num. of pre-treatment coefs
numPostPeriods = 10, # num. of post-treatment coefs
Mbarvec = seq(0.5, 2.5, by = 0.5) # values of Mbar
)Conditional parallel trends
Using did we can also estimate a model in which the trends are only parallel conditional on covariates by specifying the xformla argument.
Code
## Conditional pre-test
set.seed(123)
did_data$genre <- as.factor(did_data$genre)
conditional_gt_att <- att_gt(
yname = "log_streams",
tname = "period",
idname = "id",
gname = "G",
xformla = ~genre,
data = did_data
)
ggdid(conditional_gt_att)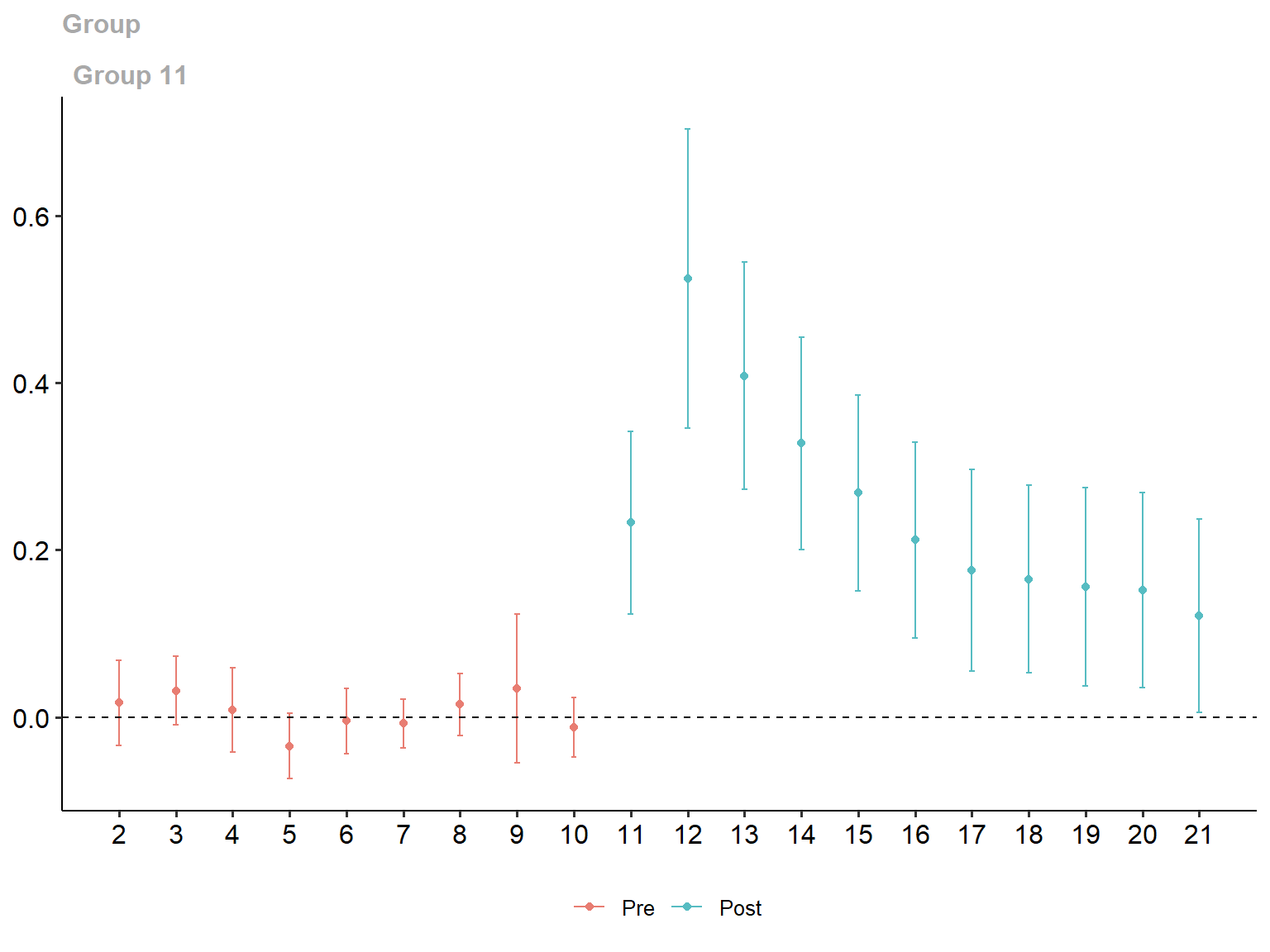
Code
summary(conditional_gt_att)
Call:
att_gt(yname = "log_streams", tname = "period", idname = "id",
gname = "G", xformla = ~genre, data = did_data)
Reference: Callaway, Brantly and Pedro H.C. Sant'Anna. "Difference-in-Differences with Multiple Time Periods." Journal of Econometrics, Vol. 225, No. 2, pp. 200-230, 2021. <https://doi.org/10.1016/j.jeconom.2020.12.001>, <https://arxiv.org/abs/1803.09015>
Group-Time Average Treatment Effects:
Group Time ATT(g,t) Std. Error [95% Simult. Conf. Band]
11 2 0.0169 0.0182 -0.0339 0.0677
11 3 0.0316 0.0147 -0.0093 0.0726
11 4 0.0085 0.0181 -0.0420 0.0591
11 5 -0.0346 0.0140 -0.0737 0.0046
11 6 -0.0047 0.0139 -0.0435 0.0341
11 7 -0.0075 0.0104 -0.0366 0.0215
11 8 0.0149 0.0131 -0.0217 0.0515
11 9 0.0345 0.0319 -0.0545 0.1235
11 10 -0.0123 0.0126 -0.0475 0.0229
11 11 0.2329 0.0392 0.1236 0.3423 *
11 12 0.5249 0.0642 0.3459 0.7038 *
11 13 0.4086 0.0487 0.2727 0.5445 *
11 14 0.3276 0.0456 0.2003 0.4548 *
11 15 0.2684 0.0421 0.1509 0.3859 *
11 16 0.2119 0.0419 0.0949 0.3289 *
11 17 0.1758 0.0433 0.0550 0.2965 *
11 18 0.1651 0.0403 0.0528 0.2774 *
11 19 0.1561 0.0426 0.0374 0.2748 *
11 20 0.1519 0.0418 0.0354 0.2684 *
11 21 0.1211 0.0414 0.0057 0.2366 *
---
Signif. codes: `*' confidence band does not cover 0
P-value for pre-test of parallel trends assumption: 0.16937
Control Group: Never Treated, Anticipation Periods: 0
Estimation Method: Doubly RobustCode
aggte(conditional_gt_att, type = "simple")
Call:
aggte(MP = conditional_gt_att, type = "simple")
Reference: Callaway, Brantly and Pedro H.C. Sant'Anna. "Difference-in-Differences with Multiple Time Periods." Journal of Econometrics, Vol. 225, No. 2, pp. 200-230, 2021. <https://doi.org/10.1016/j.jeconom.2020.12.001>, <https://arxiv.org/abs/1803.09015>
ATT Std. Error [ 95% Conf. Int.]
0.2495 0.0382 0.1746 0.3243 *
---
Signif. codes: `*' confidence band does not cover 0
Control Group: Never Treated, Anticipation Periods: 0
Estimation Method: Doubly RobustHeterogeneous treatment effects
Heterogeneity across groups
Estimation of treatment effect heterogeneity across genres using fixest.
Code
# heterogeneity across treated units
# example genre
did_m4 <- feols(
log(streams + 1) ~ treated_post * as.factor(genre) |
song_id + week,
cluster = "song_id",
data = did_data %>% dplyr::filter(week != as.Date("2018-03-19"))
)
etable(did_m4, se = "cluster")Heterogeneity across time
Estimation of different treatment effects for multiple post-treatment periods using fixest.
Code
# heterogeneity across time
did_data$treated_post_1 <- ifelse(did_data$week > as.Date("2018-03-19") & did_data$week <= (as.Date("2018-03-19") + 21) & did_data$treated == 1, 1, 0)
did_data$treated_post_2 <- ifelse(did_data$week > as.Date("2018-04-09") & did_data$week <= (as.Date("2018-04-09") + 21) & did_data$treated == 1, 1, 0)
did_data$treated_post_3 <- ifelse(did_data$week > as.Date("2018-04-30") & did_data$treated == 1, 1, 0)
did_m5 <- feols(
log(streams + 1) ~ treated_post_1 + treated_post_2 + treated_post_3 |
song_id + week,
cluster = "song_id",
data = did_data %>% dplyr::filter(week != as.Date("2018-03-19"))
)
etable(did_m5, se = "cluster")Again, the aggregation scheme in did can be used to get an equivalent estimator using the methodology proposed in Callaway and Sant’Anna (2021b).
Code
t_post_1 <- aggte(simple_gt_att, "dynamic", min_e = 1, max_e = 3)
t_post_2 <- aggte(simple_gt_att, "dynamic", min_e = 4, max_e = 6)
t_post_3 <- aggte(simple_gt_att, "dynamic", min_e = 7)
data.frame(
period = c("post 1", "post 2", "post 3"),
att = c(t_post_1$overall.att, t_post_2$overall.att, t_post_3$overall.att),
se = c(t_post_1$overall.se, t_post_2$overall.se, t_post_3$overall.se)
)Code
t_post_1
Call:
aggte(MP = simple_gt_att, type = "dynamic", min_e = 1, max_e = 3)
Reference: Callaway, Brantly and Pedro H.C. Sant'Anna. "Difference-in-Differences with Multiple Time Periods." Journal of Econometrics, Vol. 225, No. 2, pp. 200-230, 2021. <https://doi.org/10.1016/j.jeconom.2020.12.001>, <https://arxiv.org/abs/1803.09015>
Overall summary of ATT's based on event-study/dynamic aggregation:
ATT Std. Error [ 95% Conf. Int.]
0.441 0.0493 0.3444 0.5375 *
Dynamic Effects:
Event time Estimate Std. Error [95% Simult. Conf. Band]
1 0.5485 0.0665 0.4010 0.6960 *
2 0.4236 0.0516 0.3091 0.5382 *
3 0.3508 0.0461 0.2486 0.4530 *
---
Signif. codes: `*' confidence band does not cover 0
Control Group: Never Treated, Anticipation Periods: 0
Estimation Method: Doubly RobustMatching treated and control units
Ensuring covariate similarity using weighting (WeightIt package).
Code
library(WeightIt) # https://github.com/ngreifer/WeightIt
# prepare matching data
psm_matching <- did_data %>%
group_by(song_id) %>%
dplyr::summarise(
treated = max(treated),
genre = dplyr::first(genre),
danceability = dplyr::first(danceability),
valence = dplyr::first(valence),
duration_ms = dplyr::first(duration_ms),
major_label = dplyr::first(major_label),
playlist_follower_pre = mean(log(playlist_followers[week <= as.Date("2018-03-12")] + 1), na.rm = T),
n_playlists_pre = mean(log(n_playlists[week <= as.Date("2018-03-12")] + 1), na.rm = T),
artist_fame_pre = mean(log(artist_fame[week <= as.Date("2018-03-12")] + 1), na.rm = T),
ig_followers_pre = mean(log(ig_followers[week <= as.Date("2018-03-12")] + 1), na.rm = T),
streams_pre1 = mean(log(streams[week <= (as.Date("2018-03-19") - 7 * 1)] + 1)),
streams_pre2 = mean(log(streams[week <= (as.Date("2018-03-19") - 7 * 2)] + 1)),
streams_pre3 = mean(log(streams[week <= (as.Date("2018-03-19") - 7 * 3)] + 1)),
streams_pre4 = mean(log(streams[week <= (as.Date("2018-03-19") - 7 * 4)] + 1)),
streams_pre5 = mean(log(streams[week <= (as.Date("2018-03-19") - 7 * 5)] + 1)),
streams_pre6 = mean(log(streams[week <= (as.Date("2018-03-19") - 7 * 6)] + 1)),
streams_pre7 = mean(log(streams[week <= (as.Date("2018-03-19") - 7 * 7)] + 1)),
streams_pre8 = mean(log(streams[week <= (as.Date("2018-03-19") - 7 * 8)] + 1)),
streams_pre9 = mean(log(streams[week <= (as.Date("2018-03-19") - 7 * 9)] + 1)),
streams_pre10 = mean(log(streams[week <= (as.Date("2018-03-19") - 7 * 10)] + 1))
) %>%
as.data.frame()
head(psm_matching)Code
# estimate treatment propensity model
w_out <- weightit(
treated ~
streams_pre1 + streams_pre5 + streams_pre10 + danceability + valence +
duration_ms + playlist_follower_pre + n_playlists_pre,
data = psm_matching, estimand = "ATT", method = "ps", include.obj = T, link = "logit"
)
# results of logit model
summary(w_out$obj)
Call:
NULL
Deviance Residuals:
Min 1Q Median 3Q Max
-1.1637 -0.6905 -0.5036 -0.3317 2.4753
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -1.65606 0.17935 -9.234 < 0.0000000000000002 ***
streams_pre1 -4.71950 2.95376 -1.598 0.11124
streams_pre5 6.61084 3.66222 1.805 0.07215 .
streams_pre10 -1.94982 1.22974 -1.586 0.11399
danceability -0.49882 0.17540 -2.844 0.00479 **
valence 0.51725 0.18988 2.724 0.00686 **
duration_ms -0.04518 0.17558 -0.257 0.79714
playlist_follower_pre -0.62589 0.37265 -1.680 0.09417 .
n_playlists_pre 0.83886 0.40266 2.083 0.03815 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasibinomial family taken to be 1.025317)
Null deviance: 273.37 on 283 degrees of freedom
Residual deviance: 251.21 on 275 degrees of freedom
AIC: NA
Number of Fisher Scoring iterations: 5Assessing weights and covariate balance
Code
# matching summary
summary(w_out) Summary of weights
- Weight ranges:
Min Max
treated 1.0000 || 1.0000
control 0.0042 |--------------------------| 0.9682
- Units with 5 most extreme weights by group:
37 33 19 16 2
treated 1 1 1 1 1
120 123 70 132 254
control 0.8566 0.8641 0.9123 0.9475 0.9682
- Weight statistics:
Coef of Var MAD Entropy # Zeros
treated 0.000 0.000 -0.000 0
control 0.805 0.593 0.271 0
- Effective Sample Sizes:
Control Treated
Unweighted 231. 53
Weighted 140.4 53Code
hist(w_out$weights)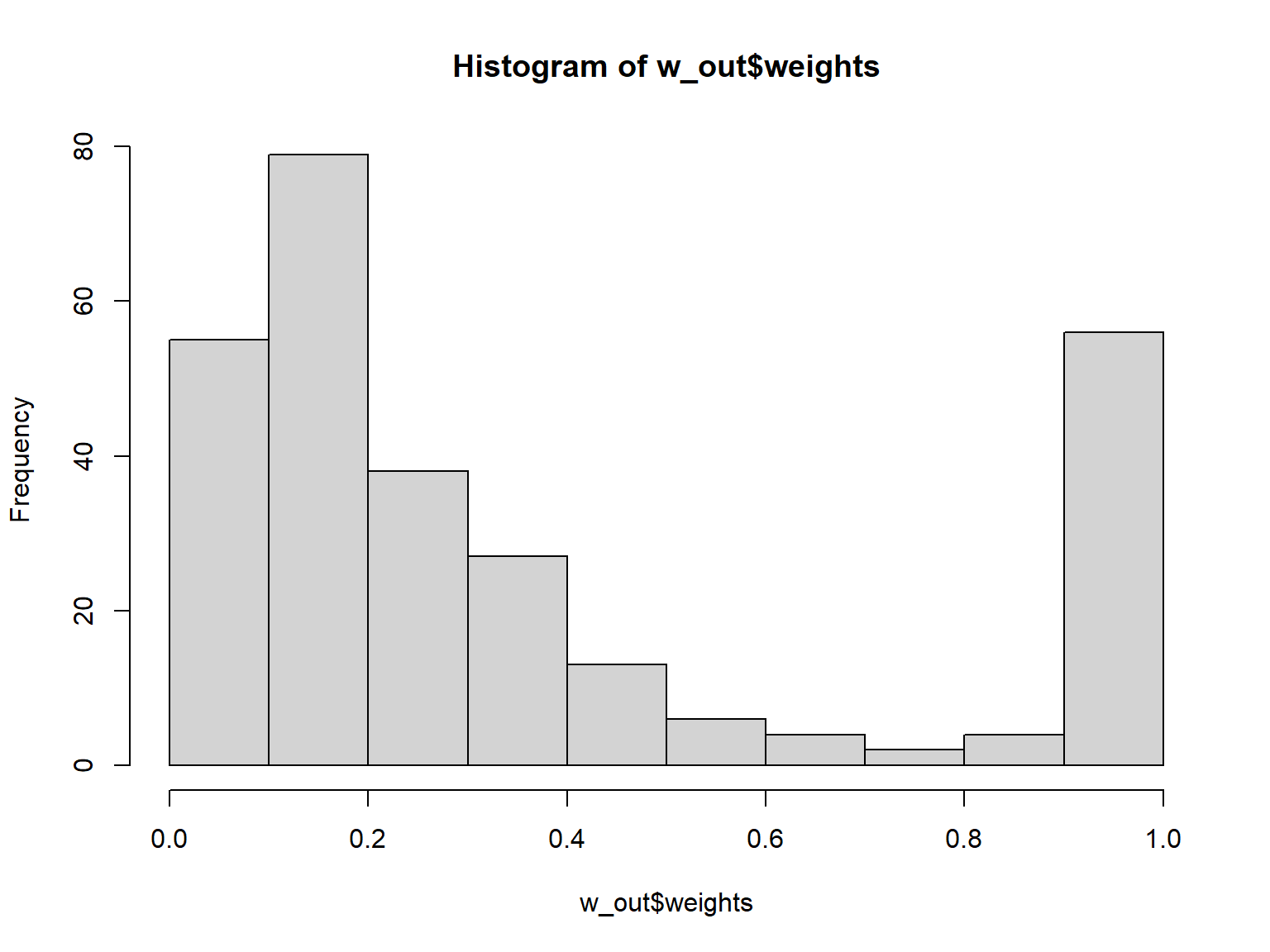
Code
# assess covariate balance
library(cobalt) # https://ngreifer.github.io/cobalt/
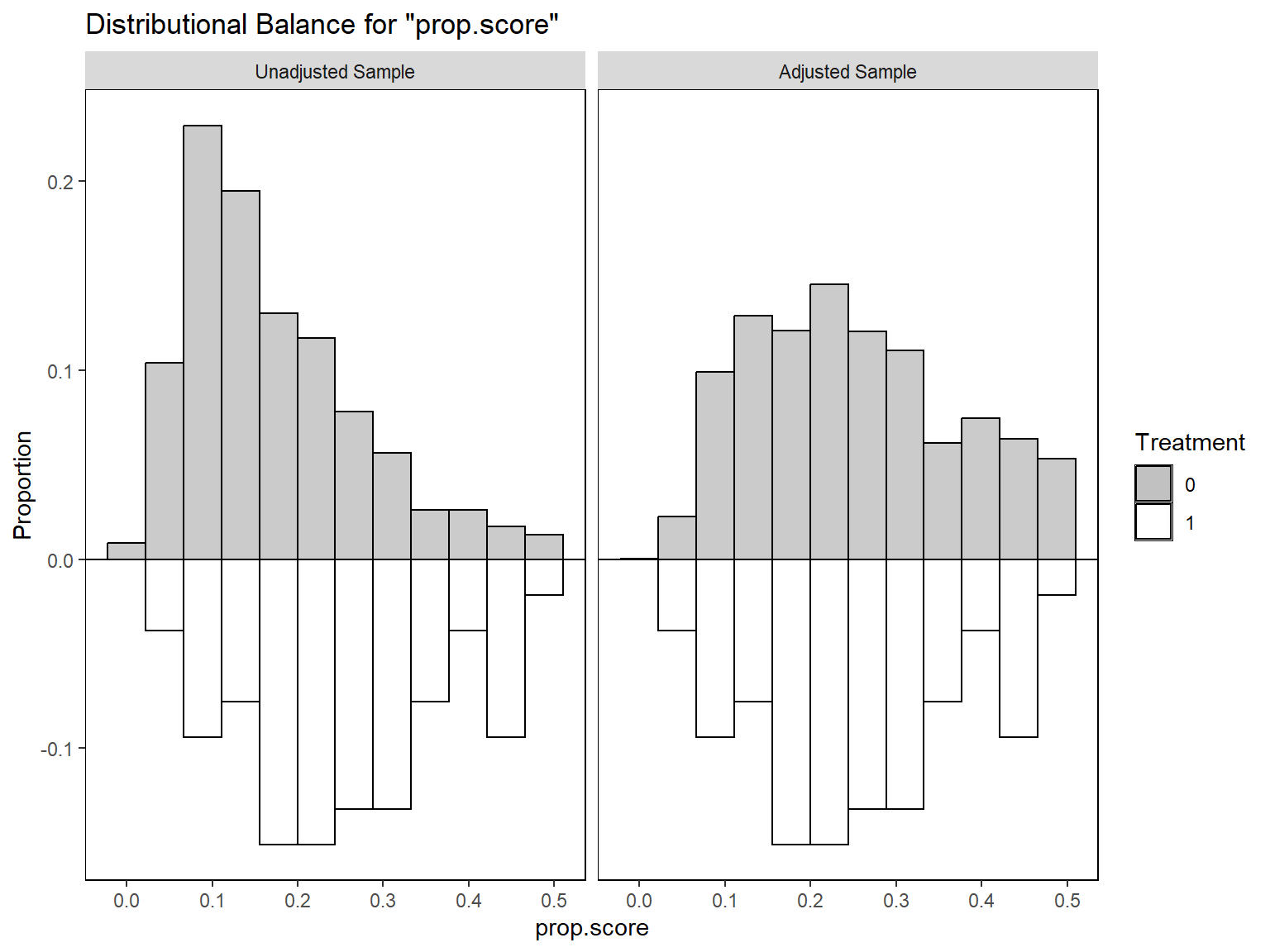
bal.tab(w_out, stats = c("m", "v"), thresholds = c(m = .05))Balance Measures
Type Diff.Adj M.Threshold V.Ratio.Adj
prop.score Distance -0.0361 Balanced, <0.05 0.8680
streams_pre1 Contin. -0.0229 Balanced, <0.05 3.3904
streams_pre5 Contin. -0.0215 Balanced, <0.05 3.4059
streams_pre10 Contin. -0.0185 Balanced, <0.05 3.2658
danceability Contin. 0.0358 Balanced, <0.05 1.1257
valence Contin. -0.0067 Balanced, <0.05 1.0781
duration_ms Contin. 0.0211 Balanced, <0.05 0.8281
playlist_follower_pre Contin. 0.0106 Balanced, <0.05 2.1967
n_playlists_pre Contin. 0.0186 Balanced, <0.05 1.4048
Balance tally for mean differences
count
Balanced, <0.05 9
Not Balanced, >0.05 0
Variable with the greatest mean difference
Variable Diff.Adj M.Threshold
danceability 0.0358 Balanced, <0.05
Effective sample sizes
Control Treated
Unadjusted 231. 53
Adjusted 140.4 53Code
bal.plot(w_out,
var.name = "prop.score", which = "both",
type = "histogram", mirror = T, colors = c("grey", "white")
)
Code
love.plot(w_out,
var.order = "unadjusted",
line = TRUE,
threshold = .1,
colors = c("darkgrey", "black")
)
Estimation using weights
The weights calculated in the previous step can be passed to the fixest package.
Code
# extract weights
weight_df <- data.frame(song_id = psm_matching$song_id, weight = w_out$weights)
# merge weights to data
did_data <- plyr::join(did_data, weight_df[, c("song_id", "weight")], type = "left", match = "first")
# estimate weighted DiD model
did_m6 <- feols(
log(streams + 1) ~ treated * post |
song_id + week,
cluster = "song_id",
weights = did_data[did_data$week != as.Date("2018-03-19"), "weight"],
data = did_data %>% dplyr::filter(week != as.Date("2018-03-19"))
)
# compare results with non-matched sample
etable(did_m2, did_m6, se = "cluster", headers = c("unweighted", "weighted"))Synthetic control
Estimation using generalized synthetic controls (gsynth package Xu and Liu 2021)) to ensure similarity between treated and control units.
Code
library(gsynth) # https://yiqingxu.org/packages/gsynth/
# inspect data
panelview(streams ~ treated_post, data = did_data, index = c("song_id", "week"), pre.post = TRUE, by.timing = TRUE)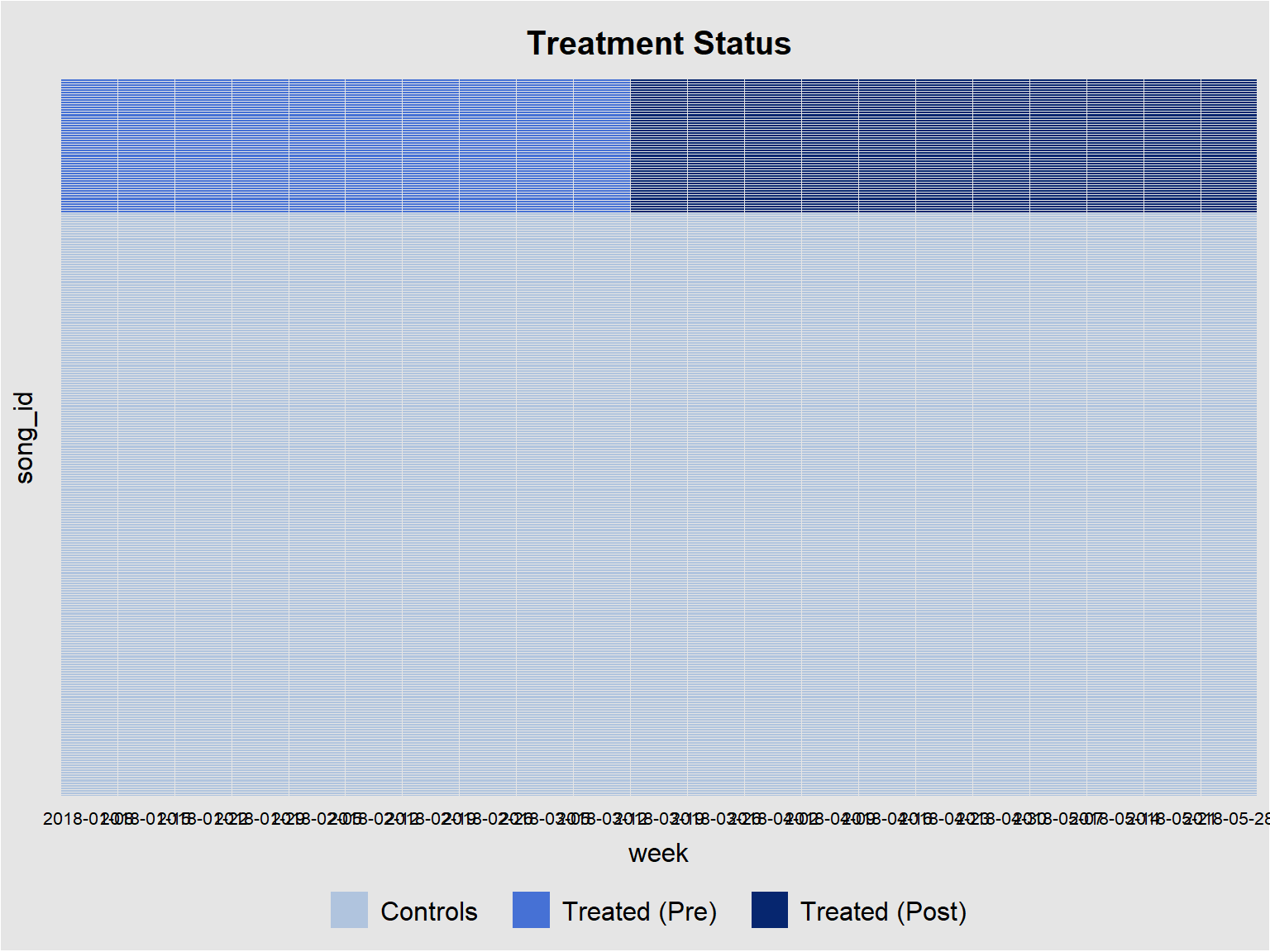
Code
did_data$log_streams <- log(did_data$streams)
did_data$log_ig_followers <- log(did_data$ig_followers + 1)
did_data$log_playlist_followers <- log(did_data$playlist_followers + 1)
did_data$log_n_postings <- log(did_data$n_postings + 1)
did_data$log_n_playlists <- log(did_data$n_playlists + 1)
# estimation
sc_m1 <- gsynth(log_streams ~ treated_post, #+
# log_ig_followers + log_playlist_followers +
# log_n_postings + log_n_playlists,
data = did_data,
# r = 0, CV = FALSE,
r = c(0, 5), CV = TRUE,
index = c("song_id", "week"), force = "two-way", se = TRUE,
inference = "parametric", nboots = 1000,
parallel = TRUE, cores = 4
)Parallel computing ...
Cross-validating ...
r = 0; sigma2 = 0.03054; IC = -3.04961; PC = 0.02896; MSPE = 0.01636
r = 1; sigma2 = 0.01090; IC = -3.64449; PC = 0.01456; MSPE = 0.01314
r = 2; sigma2 = 0.00728; IC = -3.61560; PC = 0.01255; MSPE = 0.01061*
r = 3; sigma2 = 0.00542; IC = -3.48293; PC = 0.01144; MSPE = 0.01476
r = 4; sigma2 = 0.00421; IC = -3.30913; PC = 0.01054; MSPE = 0.02762
r = 5; sigma2 = 0.00353; IC = -3.06336; PC = 0.01021; MSPE = 0.03692
r* = 2
Simulating errors ...
Bootstrapping ...
Code
# ATT
cumu_eff <- cumuEff(sc_m1, cumu = TRUE, id = NULL, period = c(0, 10)) # cumulative ATT
plot(sc_m1) # effects plot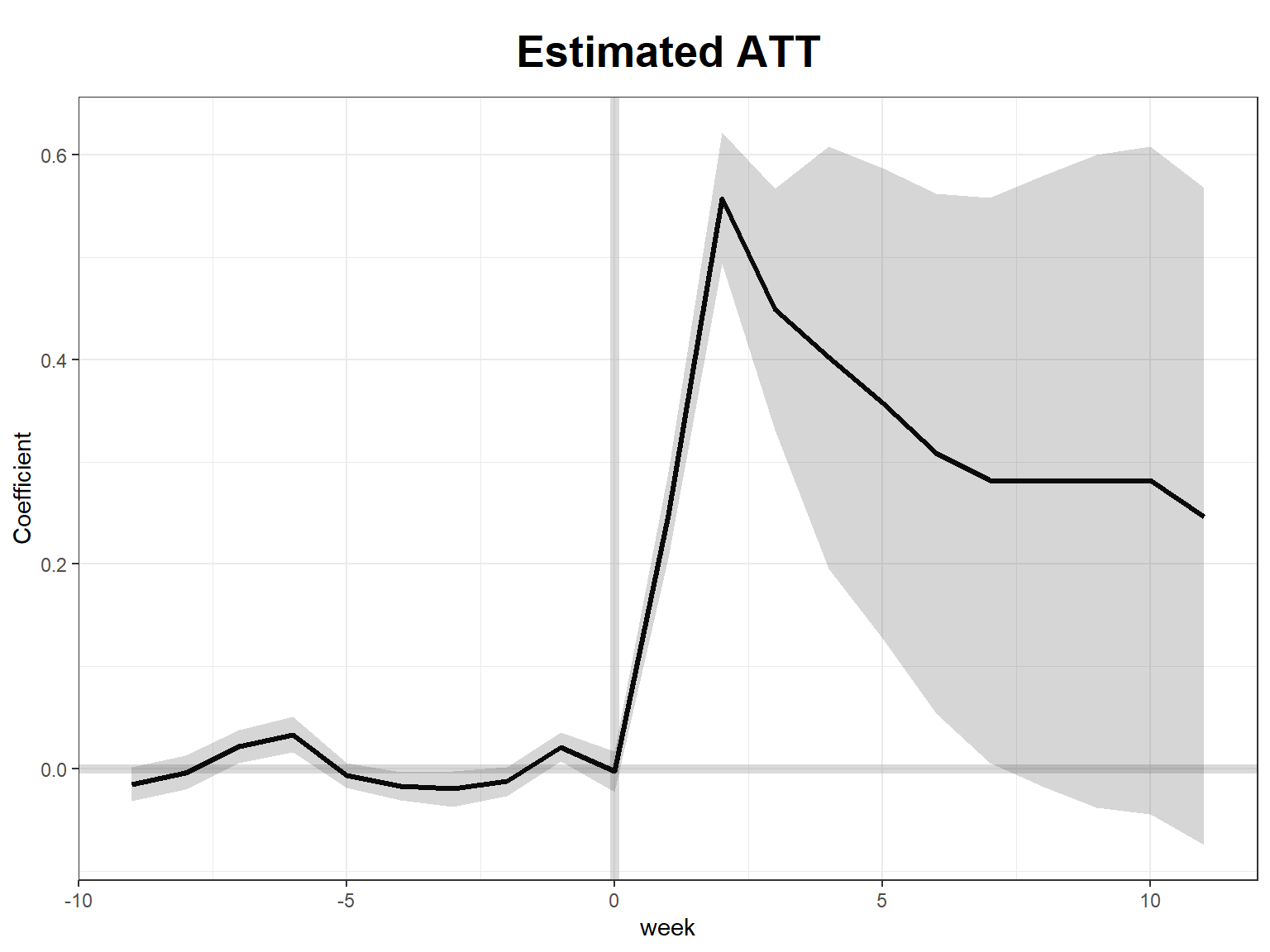
Code
plot(sc_m1, type = "raw") # incl raw data
Code
plot(sc_m1, type = "gap", id = 3, main = "Example song")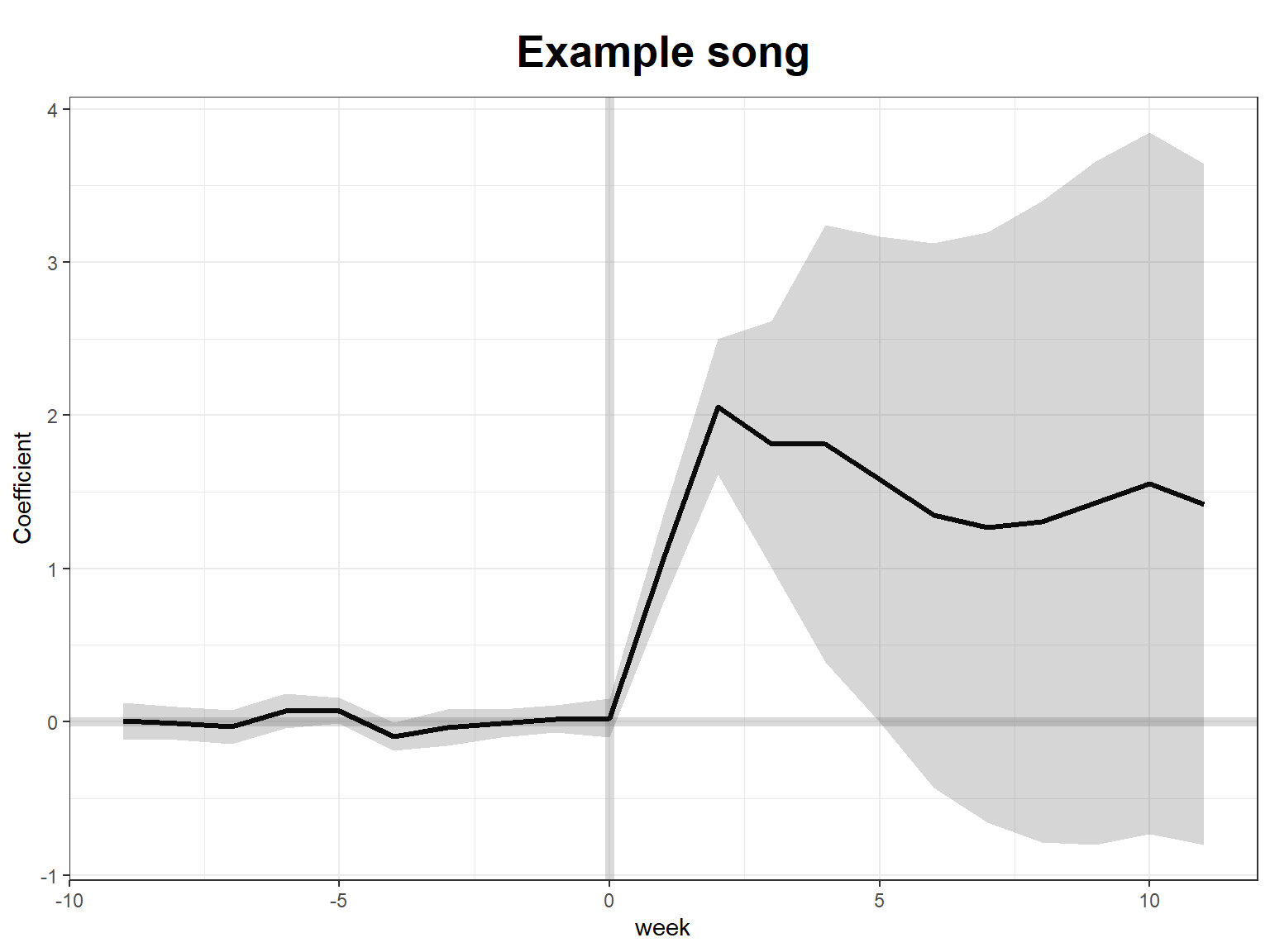
ATT and CI per period:
Code
sc_m1$est.att ATT S.E. CI.lower CI.upper p.value
-9 -0.014939001 0.009835151 -0.034215543 0.0043375411 0.128777691495282509
-8 -0.003669063 0.009118334 -0.021540670 0.0142025425 0.687402103844154233
-7 0.021725473 0.008478780 0.005107370 0.0383435769 0.010397104371477450
-6 0.033490588 0.009877606 0.014130836 0.0528503397 0.000697507019014054
-5 -0.006275912 0.007080434 -0.020153308 0.0076014844 0.375416100662471663
-4 -0.017000624 0.006890679 -0.030506107 -0.0034951408 0.013617757954593745
-3 -0.019701873 0.009574440 -0.038467432 -0.0009363152 0.039613447849829786
-2 -0.012366284 0.007451635 -0.026971220 0.0022386521 0.097007087157054306
-1 0.021280904 0.007589369 0.006406013 0.0361557941 0.005046647266487847
0 -0.002544207 0.009757656 -0.021668862 0.0165804482 0.794293362266164982
1 0.247998960 0.022163285 0.204559719 0.2914382009 0.000000000000000000
2 0.557737597 0.033714332 0.491658720 0.6238164743 0.000000000000000000
3 0.448963532 0.064991561 0.321582413 0.5763446521 0.000000000004914291
4 0.401958108 0.111697584 0.183034865 0.6208813505 0.000319899810204527
5 0.357872236 0.124435205 0.113983717 0.6017607553 0.004027847000901419
6 0.308301033 0.138397423 0.037047068 0.5795549978 0.025903857310217937
7 0.282250622 0.151077118 -0.013855089 0.5783563320 0.061726498538240859
8 0.281267992 0.164722091 -0.041581375 0.6041173577 0.087723497357681257
9 0.281026795 0.178466314 -0.068760753 0.6308143427 0.115331026171494599
10 0.281880510 0.182777276 -0.076356367 0.6401173879 0.123023145010136004
11 0.246654834 0.178396214 -0.102995321 0.5963049895 0.166780277820889333
n.Treated
-9 0
-8 0
-7 0
-6 0
-5 0
-4 0
-3 0
-2 0
-1 0
0 0
1 53
2 53
3 53
4 53
5 53
6 53
7 53
8 53
9 53
10 53
11 53ATT
Code
sc_m1$est.avg # ATT Estimate S.E. CI.lower CI.upper p.value
ATT.avg 0.335992 0.1190547 0.102649 0.569335 0.004770071\(\beta\) coefficients from factor model
Code
sc_m1$est.beta # beta coefficients from factor modelNULLCounterfactual prediction
Code
# counterfactual prediction
plot(sc_m1, type = "counterfactual", raw = "none", xlab = "Time")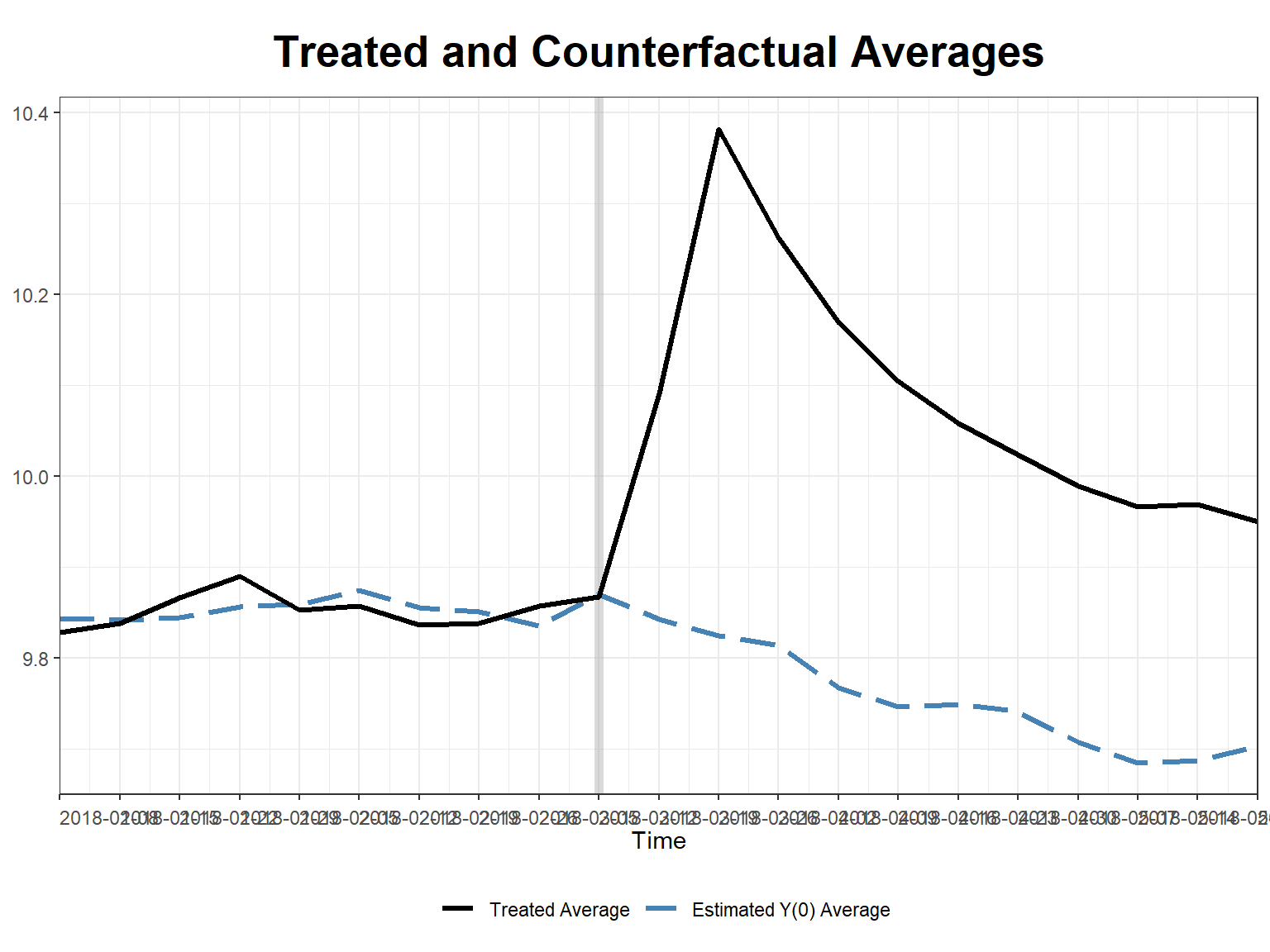
Code
plot(sc_m1, type = "counterfactual", raw = "all") # incl raw data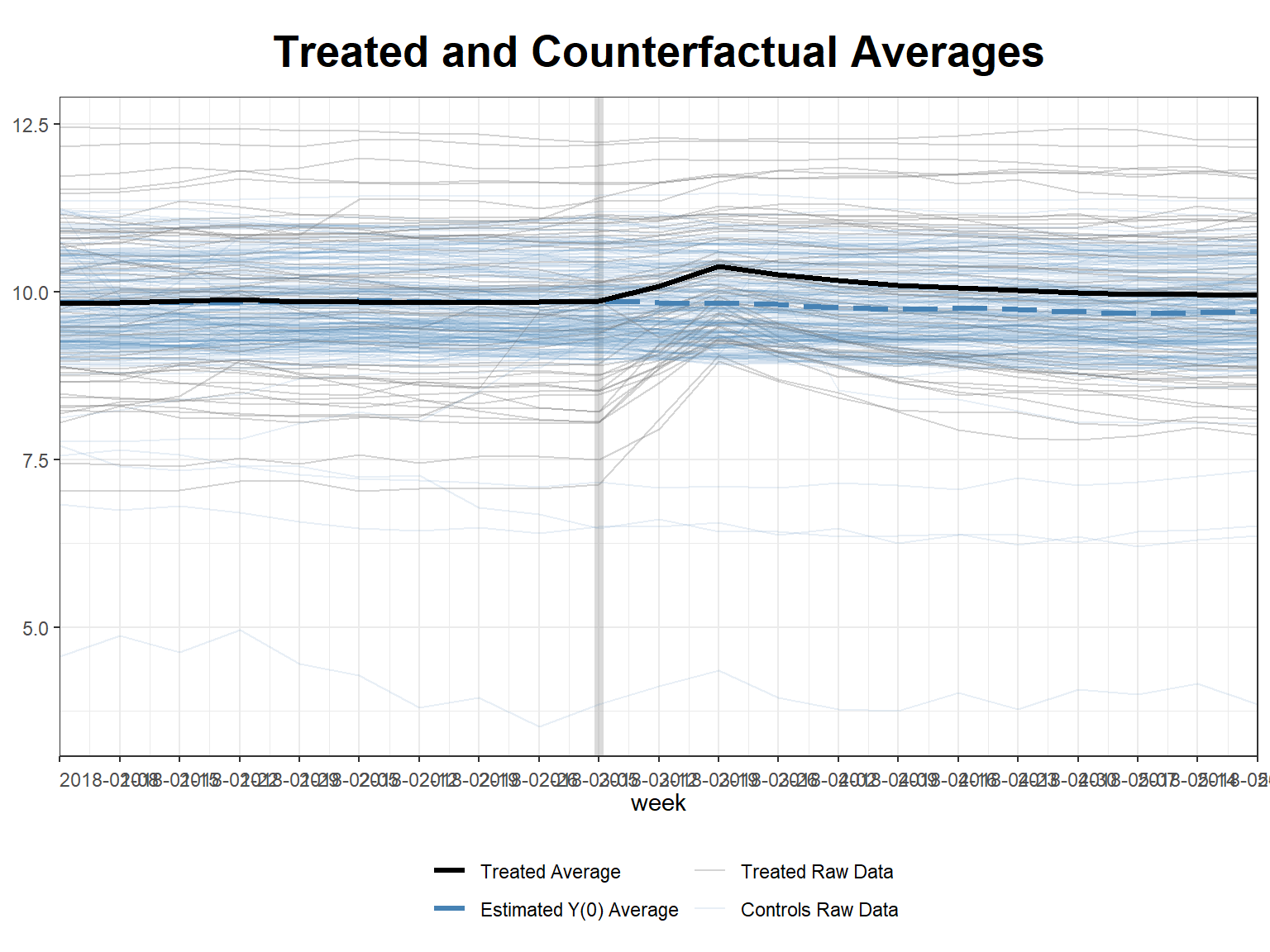
Code
plot(sc_m1, type = "counterfactual", id = 3) # for individual units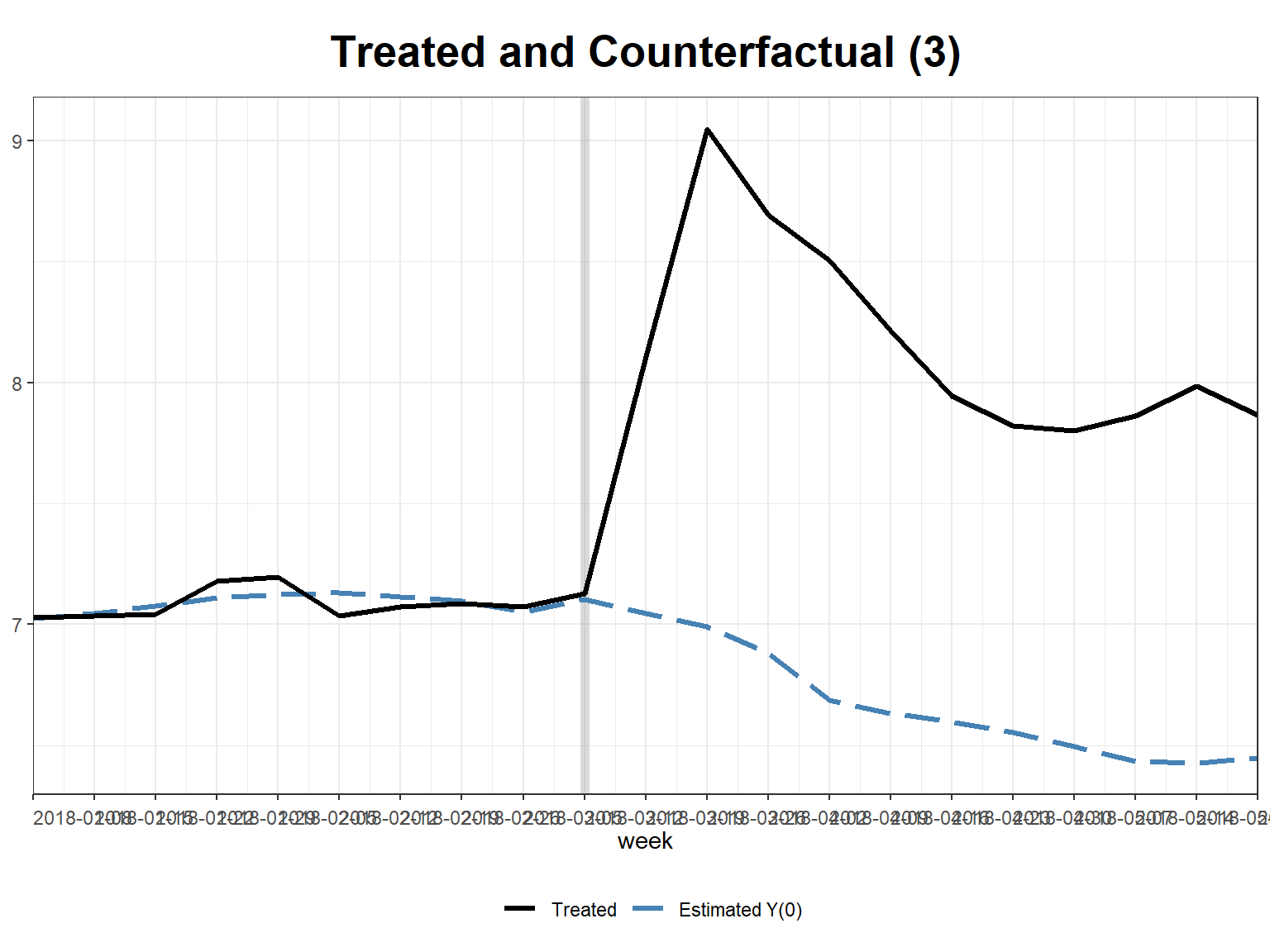
Factor model
Code
# factor model
plot(sc_m1, type = "factors", xlab = "Time")
Code
plot(sc_m1, type = "loadings")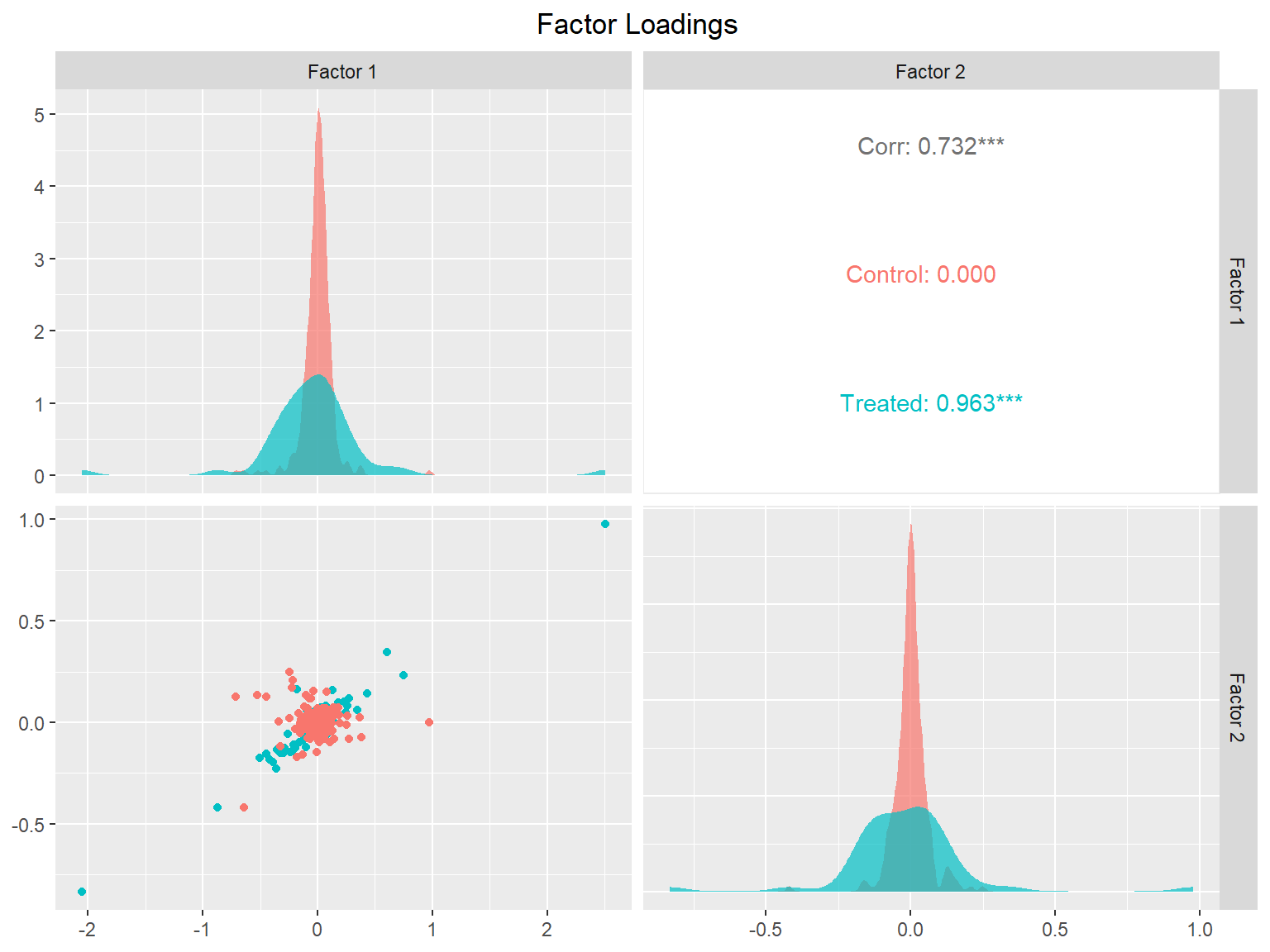
Synthetic Difference-in-Differences
Estimation using synthetic difference-in-differences proposed by Arkhangelsky et al. (2021) and implemented in the synthdid package (Arkhangelsky 2023).
Code
library(synthdid) # https://synth-inference.github.io/synthdid/articles/synthdid.html
# data pre-processing
did_data_synthdid <- did_data %>%
dplyr::select(song_id, week, streams, treated_post) %>%
dplyr::mutate(streams = log(streams)) %>%
as.data.frame()
# data setup
setup <- panel.matrices(did_data_synthdid)
# estimation
synthdid_m1 <- synthdid_estimate(setup$Y, setup$N0, setup$T0)Code
# treatment effect
plot(synthdid_m1)
Code
se <- sqrt(vcov(synthdid_m1, method = "placebo"))
sprintf("point estimate: %1.2f", synthdid_m1)[1] "point estimate: 0.27"Code
sprintf("95%% CI (%1.2f, %1.2f)", synthdid_m1 - 1.96 * se, synthdid_m1 + 1.96 * se)[1] "95% CI (0.21, 0.32)"Visualizing the results
Code
# control unit contribution
synthdid_units_plot(synthdid_m1, se.method = "placebo")
Code
# assessing parallel pre-treatment trends
plot(synthdid_m1, overlay = 1, se.method = "placebo")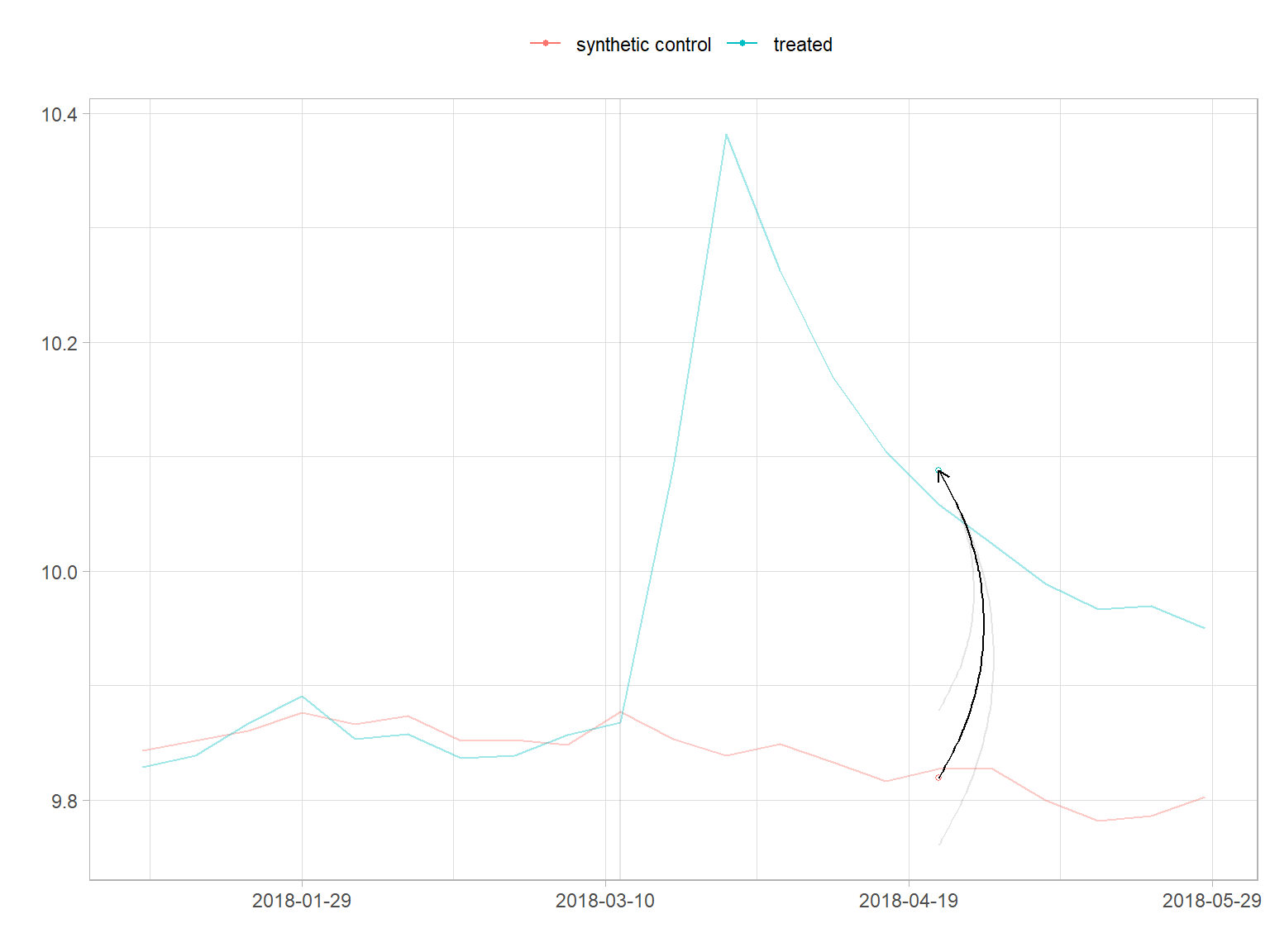
Code
plot(synthdid_m1, overlay = .8, se.method = "placebo")
Code
# compare to standard DiD estimator and synthetic control
synthdid_m2 <- did_estimate(setup$Y, setup$N0, setup$T0)
synthdid_m3 <- sc_estimate(setup$Y, setup$N0, setup$T0)
estimates <- list(synthdid_m2, synthdid_m1, synthdid_m3)
names(estimates) <- c("Diff-in-Diff", "Synthetic Diff-in-Diff", "Synthetic Control")
print(unlist(estimates)) Diff-in-Diff Synthetic Diff-in-Diff Synthetic Control
0.2836426 0.2683169 0.2908847 Time-varying treatment effects
Staggered adoption
Visualization of the panel structure using panelView.
Code
# staggered adoption
# https://yiqingxu.org/packages/gsynth/articles/tutorial.html
# upcoming package: https://github.com/zachporreca/staggered_adoption_synthdid
# load data
did_data_staggered <- fread("https://raw.githubusercontent.com/WU-RDS/RMA2022/main/data/did_data_staggered.csv")
did_data_staggered$song_id <- as.character(did_data_staggered$song_id)
did_data_staggered$week <- as.Date(did_data_staggered$week)
# data preparation
did_data_staggered$log_streams <- log(did_data_staggered$streams + 1)
did_data_staggered$log_song_age <- log(did_data_staggered$song_age + 1)
did_data_staggered$log_n_playlists <- log(did_data_staggered$n_playlists + 1)
did_data_staggered$log_playlist_followers <- log(did_data_staggered$playlist_followers + 1)
did_data_staggered$log_n_postings <- log(did_data_staggered$n_postings + 1)
# inspect data
panelview(log_streams ~ treated_post, data = did_data_staggered, index = c("song_id", "week"), pre.post = TRUE, by.timing = TRUE)
Code
panelview(log_streams ~ treated_post, data = did_data_staggered, index = c("song_id", "week"), type = "outcome")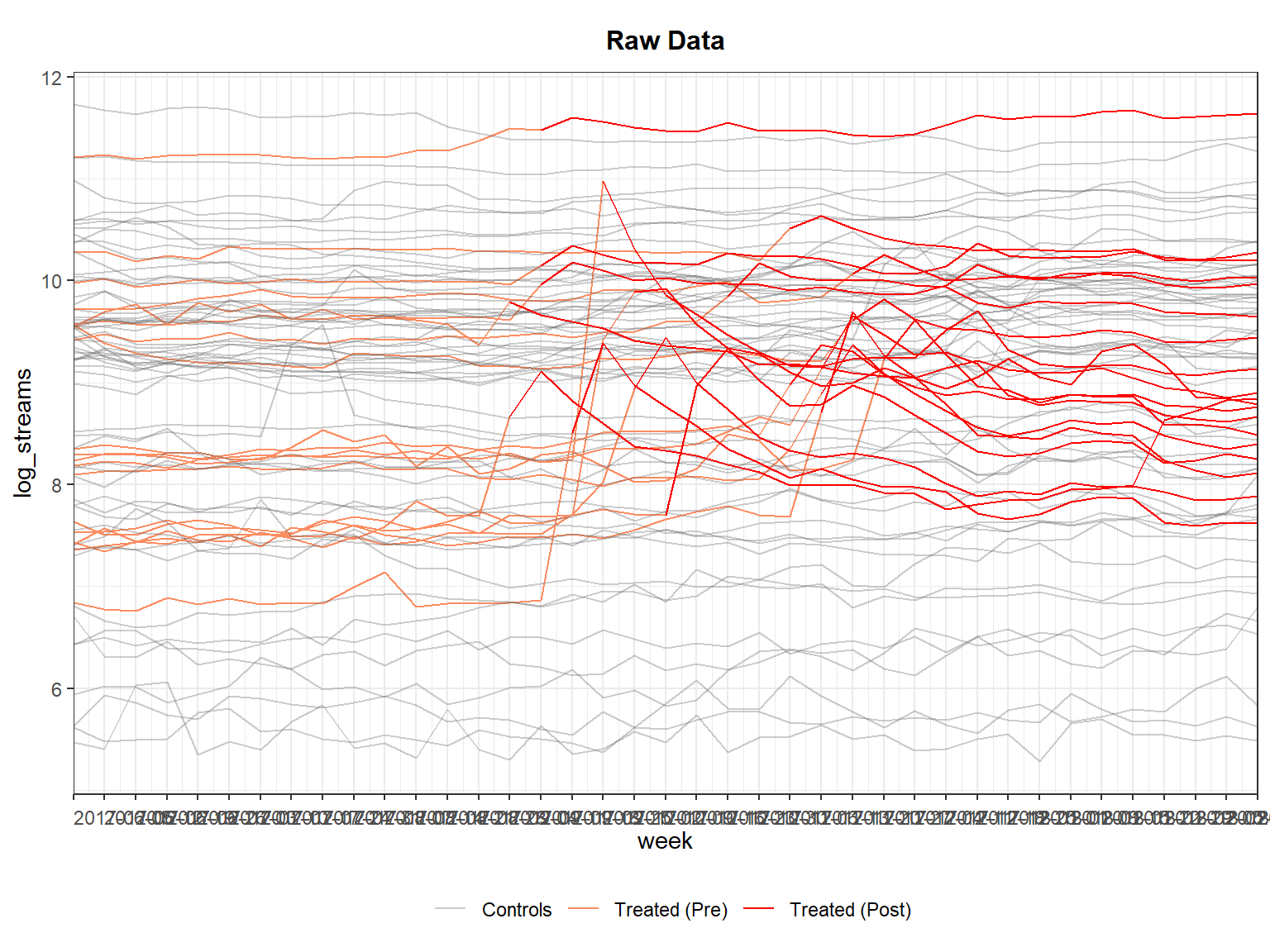
Code
panelview(log_streams ~ treated_post, data = did_data_staggered, index = c("song_id", "week"), type = "outcome", main = "Number of weekly streams", by.group = TRUE)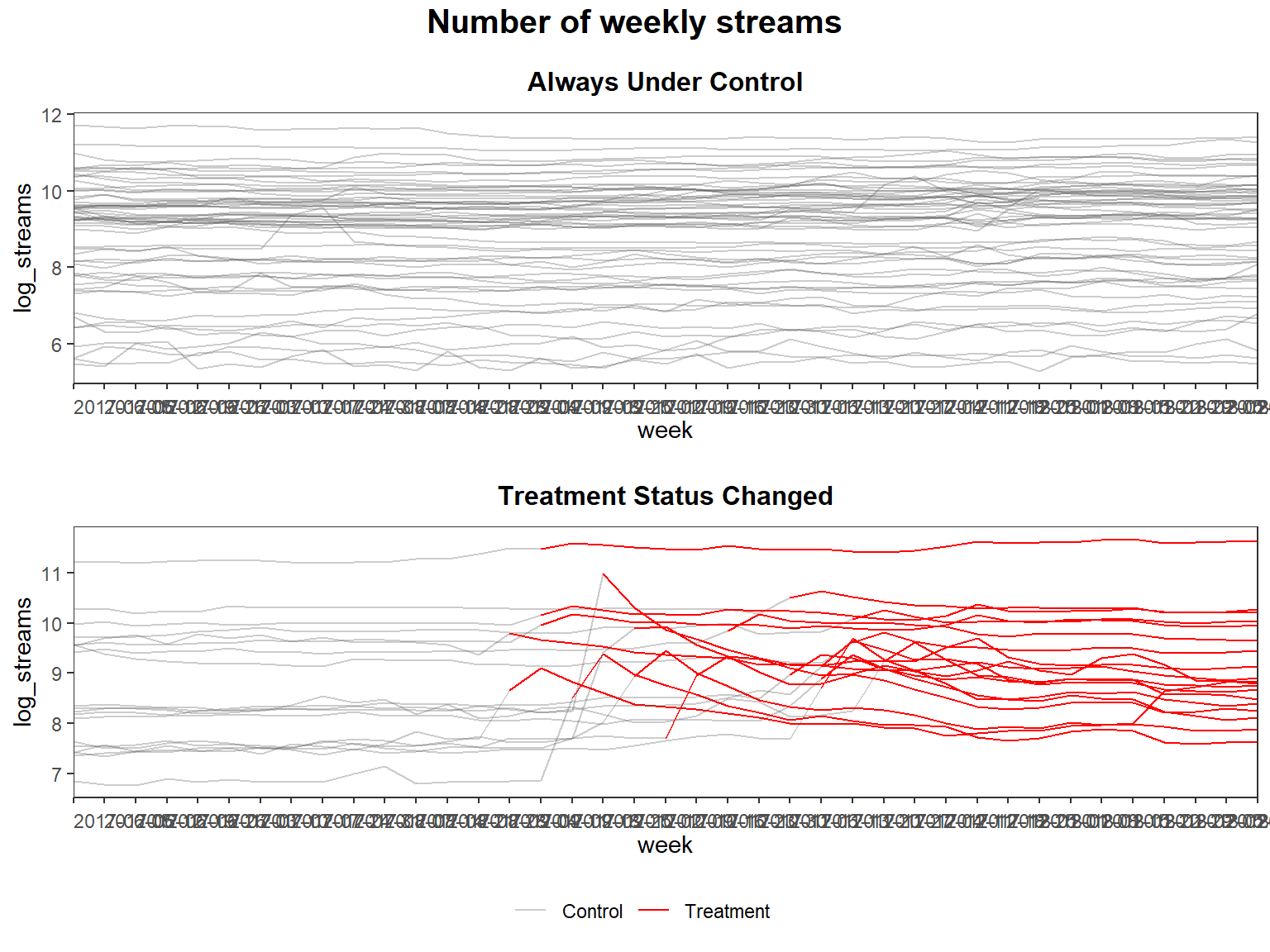
Multiple packages implement estimators appropriate for staggered adoption. Among them are gsynth, did, and fixest (latter not shown here, see ?sunab). There is also forthcoming package to extend synthdid.
Synthetic control
Estimation using gsynth.
Code
# run model
stag_m1 <- gsynth(log_streams ~ treated_post + log_song_age + log_n_playlists + log_playlist_followers + log_n_postings,
data = did_data_staggered, index = c("song_id", "week"),
se = TRUE, inference = "parametric",
r = c(0, 5), CV = TRUE, force = "two-way",
nboots = 1000, seed = 02139
)Parallel computing ...
Cross-validating ...
r = 0; sigma2 = 0.01479; IC = -3.87432; PC = 0.01414; MSPE = 0.01824
r = 1; sigma2 = 0.00835; IC = -4.12631; PC = 0.00939; MSPE = 0.01593*
r = 2; sigma2 = 0.00721; IC = -3.96033; PC = 0.00933; MSPE = 0.01658
r = 3; sigma2 = 0.00632; IC = -3.78512; PC = 0.00927; MSPE = 0.01738
r = 4; sigma2 = 0.00543; IC = -3.63799; PC = 0.00889; MSPE = 0.01850
r = 5; sigma2 = 0.00476; IC = -3.47679; PC = 0.00862; MSPE = 0.01968
r* = 1
Simulating errors ...
Bootstrapping ...
Code
# ATT
cumu_eff <- cumuEff(stag_m1, cumu = TRUE, id = NULL, period = c(0, 10)) # cumulative ATT
cumu_eff$catt
[1] 0.07190787 0.72201771 1.71260141 2.48563521 3.05755892 3.51060998
[7] 3.84717027 4.13102323 4.37933938 4.59954027 4.74789137
$est.catt
CATT S.E. CI.lower CI.upper p.value
0 0.07190787 0.02003151 0.03992271 0.1194863 0
1 0.72201771 0.04301672 0.65409399 0.8208971 0
2 1.71260141 0.06983871 1.59990247 1.8725702 0
3 2.48563521 0.10200752 2.31775373 2.7213509 0
4 3.05755892 0.13773130 2.82256342 3.3661126 0
5 3.51060998 0.17779308 3.20168824 3.9084697 0
6 3.84717027 0.22184775 3.47242065 4.3324431 0
7 4.13102323 0.26962386 3.65590487 4.7219944 0
8 4.37933938 0.31951904 3.81504646 5.0588090 0
9 4.59954027 0.37444657 3.92248425 5.4222747 0
10 4.74789137 0.43547676 3.96206121 5.6938628 0Code
plot(stag_m1) # effects plot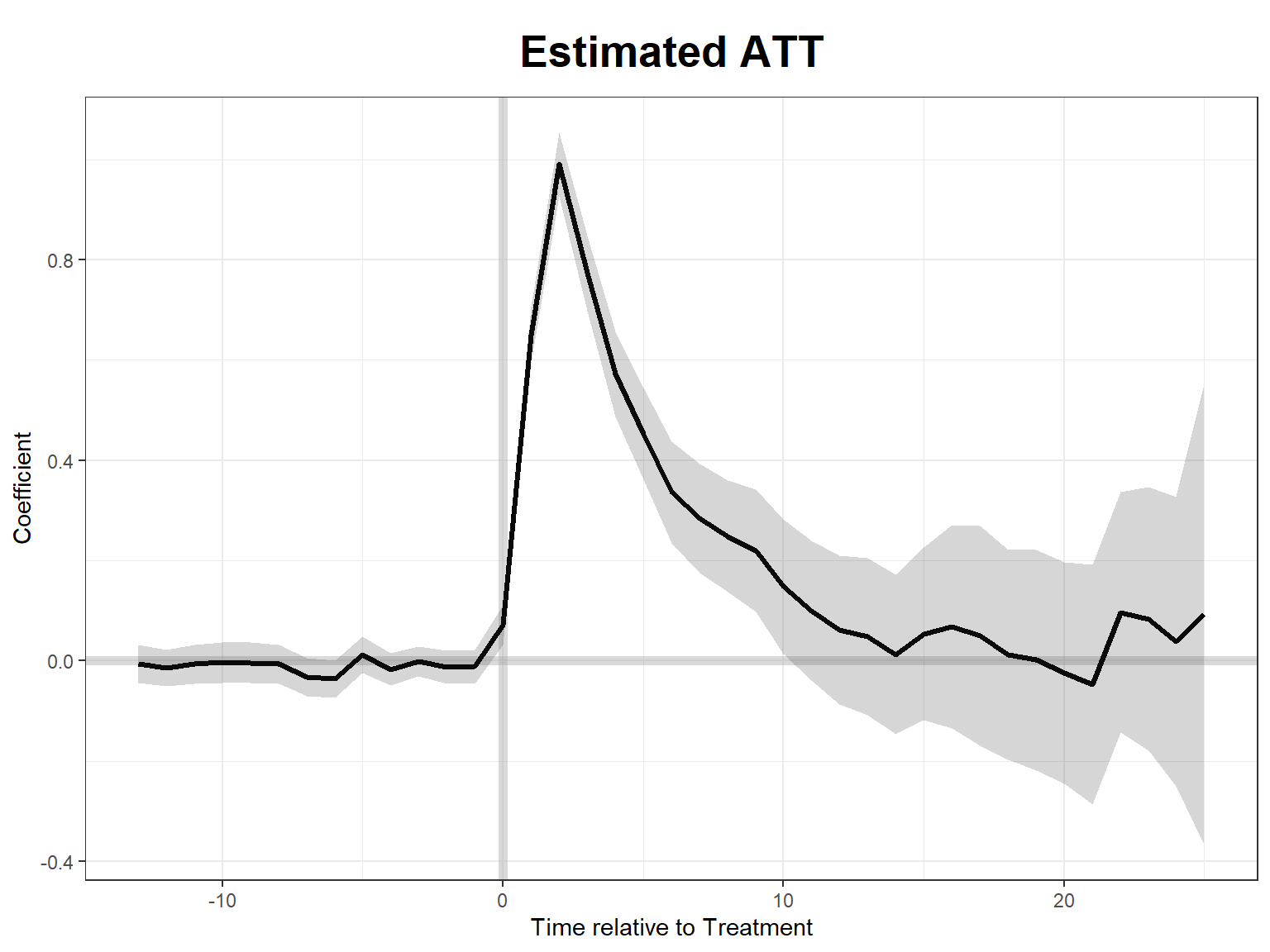
Code
plot(stag_m1, type = "gap")
Code
plot(stag_m1, type = "gap", id = 79, main = "Example song")
ATT and CI per period
Code
stag_m1$est.att # ATT and CI per period ATT S.E. CI.lower CI.upper p.value
-13 -0.0054054341 0.01927440 -0.04318256 0.032371694 0.77913511178311090
-12 -0.0138774521 0.01843034 -0.05000025 0.022245344 0.45146919840641608
-11 -0.0067336063 0.01962561 -0.04519909 0.031731877 0.73152091468251035
-10 -0.0033576945 0.02059757 -0.04372818 0.037012796 0.87050731178397744
-9 -0.0036965022 0.02036289 -0.04360704 0.036214032 0.85595056429849214
-8 -0.0065909945 0.01965459 -0.04511328 0.031931291 0.73736766963486344
-7 -0.0323289393 0.01939282 -0.07033817 0.005680290 0.09550304970414913
-6 -0.0364515566 0.01912374 -0.07393340 0.001030286 0.05663862295093280
-5 0.0123884296 0.01872200 -0.02430603 0.049082885 0.50816079660724012
-4 -0.0172533778 0.01650275 -0.04959817 0.015091417 0.29579884548013347
-3 -0.0004454345 0.01508285 -0.03000727 0.029116401 0.97643988147333327
-2 -0.0124497951 0.01690927 -0.04559135 0.020691756 0.46156603216607728
-1 -0.0115372520 0.01698733 -0.04483181 0.021757302 0.49703137286526688
0 0.0719078706 0.02003151 0.03264683 0.111168909 0.00033101069001717
1 0.6501098402 0.02864717 0.59396242 0.706257261 0.00000000000000000
2 0.9905837035 0.03208610 0.92769610 1.053471311 0.00000000000000000
3 0.7730337980 0.03857064 0.69743674 0.848630859 0.00000000000000000
4 0.5719237115 0.04324208 0.48717080 0.656676628 0.00000000000000000
5 0.4530510576 0.04702741 0.36087904 0.545223080 0.00000000000000000
6 0.3365602893 0.05174525 0.23514146 0.437979121 0.00000000007811973
7 0.2838529600 0.05565152 0.17477798 0.392927942 0.00000033868137450
8 0.2483161483 0.05662558 0.13733204 0.359300254 0.00001158638863741
9 0.2202008899 0.06236095 0.09797568 0.342426104 0.00041388184796221
10 0.1483510977 0.06859961 0.01389834 0.282803857 0.03057466523178398
11 0.0996620908 0.07098555 -0.03946702 0.238791206 0.16032563330680683
12 0.0617311668 0.07592444 -0.08707801 0.210540339 0.41618335658991956
13 0.0485301714 0.07982027 -0.10791467 0.204975017 0.54319204471373106
14 0.0126136638 0.08103570 -0.14621339 0.171440722 0.87630446794065309
15 0.0538661163 0.08791188 -0.11843800 0.226170236 0.54005586340453293
16 0.0677489377 0.10328158 -0.13467924 0.270177113 0.51184766163355100
17 0.0496789910 0.11183431 -0.16951223 0.268870209 0.65688382980198634
18 0.0116951540 0.10677328 -0.19757662 0.220966930 0.91278007049659315
19 0.0014565251 0.11210216 -0.21825967 0.221172722 0.98963350751376122
20 -0.0236014674 0.11277651 -0.24463936 0.197436427 0.83423243404798764
21 -0.0479777209 0.12199573 -0.28708496 0.191129521 0.69411729322338056
22 0.0967723399 0.12262157 -0.14356153 0.337106208 0.42999800925794518
23 0.0836290245 0.13418156 -0.17936199 0.346620044 0.53311844165248390
24 0.0385864510 0.14700569 -0.24953941 0.326712314 0.79294932185020772
25 0.0928196596 0.23493023 -0.36763513 0.553274452 0.69277309336814086
n.Treated
-13 0
-12 0
-11 0
-10 0
-9 0
-8 0
-7 0
-6 0
-5 0
-4 0
-3 0
-2 0
-1 0
0 0
1 19
2 19
3 19
4 19
5 19
6 19
7 19
8 19
9 19
10 19
11 19
12 19
13 19
14 18
15 16
16 13
17 11
18 11
19 10
20 10
21 9
22 7
23 6
24 5
25 2ATT
Code
stag_m1$est.avg # ATT Estimate S.E. CI.lower CI.upper p.value
ATT.avg 0.2640589 0.06225465 0.142042 0.3860758 0.00002219394\(\beta\) from factor model
Code
stag_m1$est.beta # beta coefficients form factor model beta S.E. CI.lower CI.upper
log_song_age -1.13361285 0.418112413 -1.953098123 -0.31412758
log_n_playlists -0.04535966 0.069146370 -0.180884054 0.09016474
log_playlist_followers 0.09602444 0.019785461 0.057245652 0.13480323
log_n_postings 0.01396278 0.005562191 0.003061087 0.02486448
p.value
log_song_age 0.006702737142
log_n_playlists 0.511827471407
log_playlist_followers 0.000001214342
log_n_postings 0.012062783443counterfactual prediction
Code
plot(stag_m1, type = "raw")
Code
plot(stag_m1, type = "counterfactual", id = 79) # for individual units
factor model
Code
plot(stag_m1, type = "factors", xlab = "Time")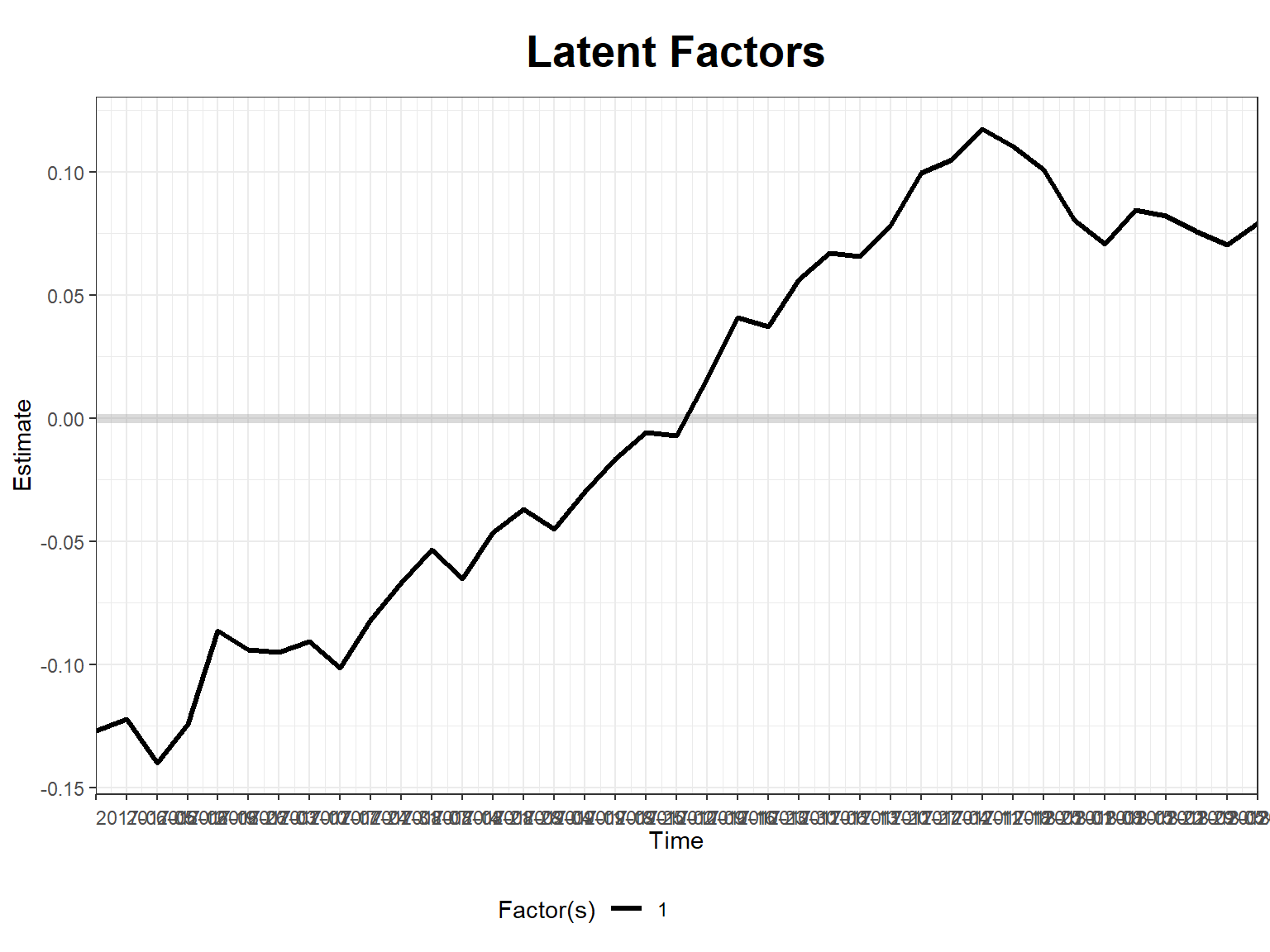
Code
plot(stag_m1, type = "loadings")Loadings for the first factor are shown...
Staggered Difference-in-Differences
Estimation using did.
Code
# staggered did
did_data_staggered$period <- as.numeric(factor(as.character(did_data_staggered$week),
levels = as.character(sort(unique(did_data_staggered$week)))
))
# add first period treated
did_data_staggered_G <- did_data_staggered |>
filter(treated == 1, week == treat_week) |>
select(song_id, G = period)
did_data_staggered <- left_join(did_data_staggered,
did_data_staggered_G,
by = "song_id"
)
did_data_staggered$G <- coalesce(did_data_staggered$G, 0)
did_data_staggered$id <- as.numeric(did_data_staggered$song_id)
set.seed(123)
# increase bootstrap for reliability!
did_stag <- att_gt(
yname = "log_streams",
tname = "period",
idname = "id",
gname = "G",
biters = 2000,
data = did_data_staggered
)Overall ATT
Code
## Full aggregation
aggte(did_stag, type = "simple", biters = 20000)
Call:
aggte(MP = did_stag, type = "simple", biters = 20000)
Reference: Callaway, Brantly and Pedro H.C. Sant'Anna. "Difference-in-Differences with Multiple Time Periods." Journal of Econometrics, Vol. 225, No. 2, pp. 200-230, 2021. <https://doi.org/10.1016/j.jeconom.2020.12.001>, <https://arxiv.org/abs/1803.09015>
ATT Std. Error [ 95% Conf. Int.]
0.3398 0.0984 0.147 0.5327 *
---
Signif. codes: `*' confidence band does not cover 0
Control Group: Never Treated, Anticipation Periods: 0
Estimation Method: Doubly RobustATT by time relative to treatment
Code
## Aggregate time relative to treatment
aggte(did_stag, type = "dynamic")
Call:
aggte(MP = did_stag, type = "dynamic")
Reference: Callaway, Brantly and Pedro H.C. Sant'Anna. "Difference-in-Differences with Multiple Time Periods." Journal of Econometrics, Vol. 225, No. 2, pp. 200-230, 2021. <https://doi.org/10.1016/j.jeconom.2020.12.001>, <https://arxiv.org/abs/1803.09015>
Overall summary of ATT's based on event-study/dynamic aggregation:
ATT Std. Error [ 95% Conf. Int.]
0.302 0.1066 0.0931 0.5108 *
Dynamic Effects:
Event time Estimate Std. Error [95% Simult. Conf. Band]
-25 0.0447 0.0138 0.0087 0.0806 *
-24 -0.0596 0.0563 -0.2063 0.0871
-23 -0.0235 0.0311 -0.1046 0.0577
-22 0.0102 0.0155 -0.0302 0.0507
-21 -0.0027 0.0214 -0.0584 0.0530
-20 0.0107 0.0149 -0.0280 0.0494
-19 0.0067 0.0207 -0.0472 0.0606
-18 0.0177 0.0260 -0.0499 0.0853
-17 0.0158 0.0197 -0.0356 0.0672
-16 0.0330 0.0188 -0.0160 0.0820
-15 -0.0272 0.0279 -0.0999 0.0455
-14 0.0104 0.0177 -0.0357 0.0564
-13 -0.0056 0.0208 -0.0597 0.0484
-12 0.0108 0.0131 -0.0234 0.0450
-11 0.0134 0.0196 -0.0377 0.0645
-10 -0.0019 0.0183 -0.0495 0.0457
-9 0.0080 0.0162 -0.0343 0.0502
-8 -0.0140 0.0171 -0.0586 0.0306
-7 0.0115 0.0178 -0.0347 0.0578
-6 0.0445 0.0130 0.0106 0.0785 *
-5 -0.0104 0.0320 -0.0937 0.0729
-4 0.0274 0.0133 -0.0073 0.0621
-3 -0.0060 0.0287 -0.0807 0.0686
-2 0.0132 0.0200 -0.0388 0.0652
-1 0.0978 0.1262 -0.2311 0.4267
0 0.6528 0.1585 0.2398 1.0658 *
1 0.9906 0.1564 0.5832 1.3981 *
2 0.7880 0.1351 0.4359 1.1400 *
3 0.6068 0.1182 0.2990 0.9147 *
4 0.4940 0.1085 0.2113 0.7767 *
5 0.3945 0.1036 0.1245 0.6645 *
6 0.3554 0.1079 0.0744 0.6365 *
7 0.3201 0.0937 0.0759 0.5643 *
8 0.2988 0.0923 0.0584 0.5391 *
9 0.2358 0.0823 0.0215 0.4501 *
10 0.1941 0.0879 -0.0349 0.4230
11 0.1692 0.0831 -0.0474 0.3857
12 0.1531 0.0769 -0.0473 0.3535
13 0.1209 0.0795 -0.0861 0.3280
14 0.1503 0.0843 -0.0693 0.3698
15 0.1738 0.0902 -0.0611 0.4087
16 0.1603 0.1078 -0.1206 0.4411
17 0.1181 0.1189 -0.1917 0.4280
18 0.1067 0.1453 -0.2718 0.4851
19 0.0772 0.1385 -0.2836 0.4379
20 0.0131 0.1459 -0.3670 0.3932
21 0.2654 0.1570 -0.1436 0.6744
22 0.2341 0.1805 -0.2362 0.7044
23 0.1844 0.2153 -0.3766 0.7455
24 0.2925 0.5287 -1.0848 1.6698
---
Signif. codes: `*' confidence band does not cover 0
Control Group: Never Treated, Anticipation Periods: 0
Estimation Method: Doubly RobustCode
ggdid(aggte(did_stag, type = "dynamic", biters = 20000))
ATT by treatment time
Code
## Aggregate total effect by treatment time
aggte(did_stag, type = "group", biters = 20000)
Call:
aggte(MP = did_stag, type = "group", biters = 20000)
Reference: Callaway, Brantly and Pedro H.C. Sant'Anna. "Difference-in-Differences with Multiple Time Periods." Journal of Econometrics, Vol. 225, No. 2, pp. 200-230, 2021. <https://doi.org/10.1016/j.jeconom.2020.12.001>, <https://arxiv.org/abs/1803.09015>
Overall summary of ATT's based on group/cohort aggregation:
ATT Std. Error [ 95% Conf. Int.]
0.3485 0.1008 0.151 0.5461 *
Group Effects:
Group Estimate Std. Error [95% Simult. Conf. Band]
15 0.1647 0.2390 -1.1764 1.5058
16 0.1304 0.0839 -0.3406 0.6014
17 1.1247 0.0169 1.0298 1.2197 *
18 0.9600 0.0164 0.8677 1.0523 *
19 -0.0924 0.1695 -1.0437 0.8589
20 0.7733 0.0146 0.6915 0.8551 *
22 0.1957 0.0167 0.1021 0.2893 *
24 0.2251 0.1168 -0.4305 0.8808
25 0.5162 0.1848 -0.5208 1.5533
26 0.2228 0.0411 -0.0081 0.4537
27 0.5879 0.0128 0.5161 0.6597 *
---
Signif. codes: `*' confidence band does not cover 0
Control Group: Never Treated, Anticipation Periods: 0
Estimation Method: Doubly RobustATT by calendar time
Code
## Aggregate total effect by calendar time
aggte(did_stag, type = "calendar", biters = 20000)
Call:
aggte(MP = did_stag, type = "calendar", biters = 20000)
Reference: Callaway, Brantly and Pedro H.C. Sant'Anna. "Difference-in-Differences with Multiple Time Periods." Journal of Econometrics, Vol. 225, No. 2, pp. 200-230, 2021. <https://doi.org/10.1016/j.jeconom.2020.12.001>, <https://arxiv.org/abs/1803.09015>
Overall summary of ATT's based on calendar time aggregation:
ATT Std. Error [ 95% Conf. Int.]
0.4059 0.1131 0.1842 0.6276 *
Time Effects:
Time Estimate Std. Error [95% Simult. Conf. Band]
15 0.7019 0.2122 0.1855 1.2182 *
16 0.4404 0.2917 -0.2694 1.1502
17 0.6635 0.2462 0.0644 1.2626 *
18 1.0019 0.4035 0.0200 1.9839 *
19 0.7178 0.2658 0.0710 1.3646 *
20 0.6379 0.2266 0.0863 1.1894 *
21 0.6142 0.2027 0.1210 1.1074 *
22 0.5491 0.1880 0.0915 1.0067 *
23 0.4428 0.1683 0.0333 0.8524 *
24 0.3142 0.1305 -0.0035 0.6319
25 0.4065 0.1231 0.1070 0.7061 *
26 0.5311 0.1496 0.1672 0.8951 *
27 0.4944 0.1190 0.2048 0.7841 *
28 0.4084 0.1183 0.1205 0.6963 *
29 0.3343 0.1018 0.0866 0.5821 *
30 0.2937 0.0841 0.0889 0.4984 *
31 0.2352 0.0751 0.0523 0.4180 *
32 0.1715 0.0794 -0.0216 0.3646
33 0.2120 0.0878 -0.0018 0.4258
34 0.2202 0.0795 0.0268 0.4136 *
35 0.2185 0.0765 0.0325 0.4046 *
36 0.1657 0.0757 -0.0184 0.3499
37 0.1462 0.0856 -0.0621 0.3545
38 0.1128 0.0901 -0.1066 0.3322
39 0.1131 0.0924 -0.1118 0.3380
---
Signif. codes: `*' confidence band does not cover 0
Control Group: Never Treated, Anticipation Periods: 0
Estimation Method: Doubly RobustComparison to two-way Fixed Effects
The TWFE estimator overestimates the treatment effect in this case
Code
# compare with standard DiD
stag_m2 <- feols(
log(streams + 1) ~ treated_post |
song_id + week,
cluster = "song_id",
data = did_data_staggered
)
etable(stag_m2, se = "cluster")Compared to Callaway and Sant’Anna (2021b):
Code
aggte(did_stag, type = "simple", biters = 20000)
Call:
aggte(MP = did_stag, type = "simple", biters = 20000)
Reference: Callaway, Brantly and Pedro H.C. Sant'Anna. "Difference-in-Differences with Multiple Time Periods." Journal of Econometrics, Vol. 225, No. 2, pp. 200-230, 2021. <https://doi.org/10.1016/j.jeconom.2020.12.001>, <https://arxiv.org/abs/1803.09015>
ATT Std. Error [ 95% Conf. Int.]
0.3398 0.097 0.1497 0.5299 *
---
Signif. codes: `*' confidence band does not cover 0
Control Group: Never Treated, Anticipation Periods: 0
Estimation Method: Doubly RobustDifference-in-Difference-in-Differences
In addition, to a canonical diff-in-diffs it adds a “counterfactual” diff-in-diff of groups similar to the original treated and control group in a unit (e.g., region) neither of them is treated.
Code
set.seed(123)
is_unsw <- c(0, 1)
is_dash <- c(0, 1)
groups <- expand.grid(is_unsw = is_unsw, is_dash = is_dash)
for (i in 1:15) {
groups <- rbind(groups, groups)
}
groups_pre <- groups |> mutate(id = seq_len(n()), is_post = 0)
groups_post <- groups_pre |> mutate(is_post = 1)
groups_df <- rbind(groups_pre, groups_post)
sample_y <- function(df) {
is_unsw <- df$is_unsw
is_dash <- df$is_dash
is_post <- df$is_post
df$y <- 10 +
2 * is_unsw +
3 * is_dash +
4 * is_unsw * is_dash +
-1.5 * is_post +
2 * is_unsw * is_post +
-3 * is_dash * is_post +
4 * is_unsw * is_dash * is_post + # <- treated with delta = 4
rnorm(nrow(df), 0, 1)
return(df)
}
panel <- sample_y(groups_df)
head(panel)Estimation of a difference-in-difference-in-differences model using fixest.
Code
model_ols <- feols(
y ~ is_unsw +
is_dash +
is_unsw * is_dash +
is_post +
is_unsw * is_post +
is_dash * is_post +
is_unsw * is_dash * is_post,
panel,
cluster = "id"
)
model_fe <- feols(
y ~ is_unsw:is_dash:is_post |
id +
is_post +
is_unsw:is_post +
is_dash:is_post,
panel
)
etable(model_ols, model_fe)Interpretation as Difference between two Diff-in-Diff models
Code
means <- panel |>
group_by(is_unsw, is_dash, is_post) |>
summarise(y = mean(y)) |>
arrange(is_post)
diffs <- means |>
group_by(is_unsw, is_dash) |>
summarize(y = diff(y))
with(
diffs,
## Diff-in-Diff for UNSW
y[is_unsw == 1 & is_dash == 1] -
y[is_unsw == 1 & is_dash == 0] -
## Counterfactual Diff-in-Diff for WU
(y[is_unsw == 0 & is_dash == 1] -
y[is_unsw == 0 & is_dash == 0])
)[1] 3.997019Bayesian structural time-series approach
Estimation of a Bayesian structural time-series model using the CausalImpact package (Brodersen et al. 2014). This method uses a predicted value, based on comparable time series and systematic components of the treated unit as the counterfactual post-treatment.
Code
library(CausalImpact) # https://google.github.io/CausalImpact/CausalImpact.html
# load data
ci_data <- fread("https://raw.githubusercontent.com/WU-RDS/RMA2022/main/data/ci_data.csv")
# data preparation
# control songs
ctrl_data <- ci_data %>%
dplyr::filter(treated == 0) %>%
as.data.frame()
ctrl_data_wide <- spread(ctrl_data[, -4], song_id, streams)
head(ctrl_data_wide)Visualization of the time series
Code
x <- ctrl_data_wide[, -1]
# treated song
treated_data <- ci_data %>%
dplyr::filter(treated == 1) %>%
as.data.frame()
y <- treated_data[, 3]
# run CI model
# create dataframe
data_ci <- cbind(y, x)
# visualize time series
matplot(data_ci, type = "l")
Estimation of the model and visualization of the results
Code
# specify number of pre and post periods
pre_period <- c(1, 38)
post_period <- c(38 + 1, 77)
# run model
ci_m1 <- CausalImpact(data_ci, pre_period, post_period)
# result summary
plot(ci_m1)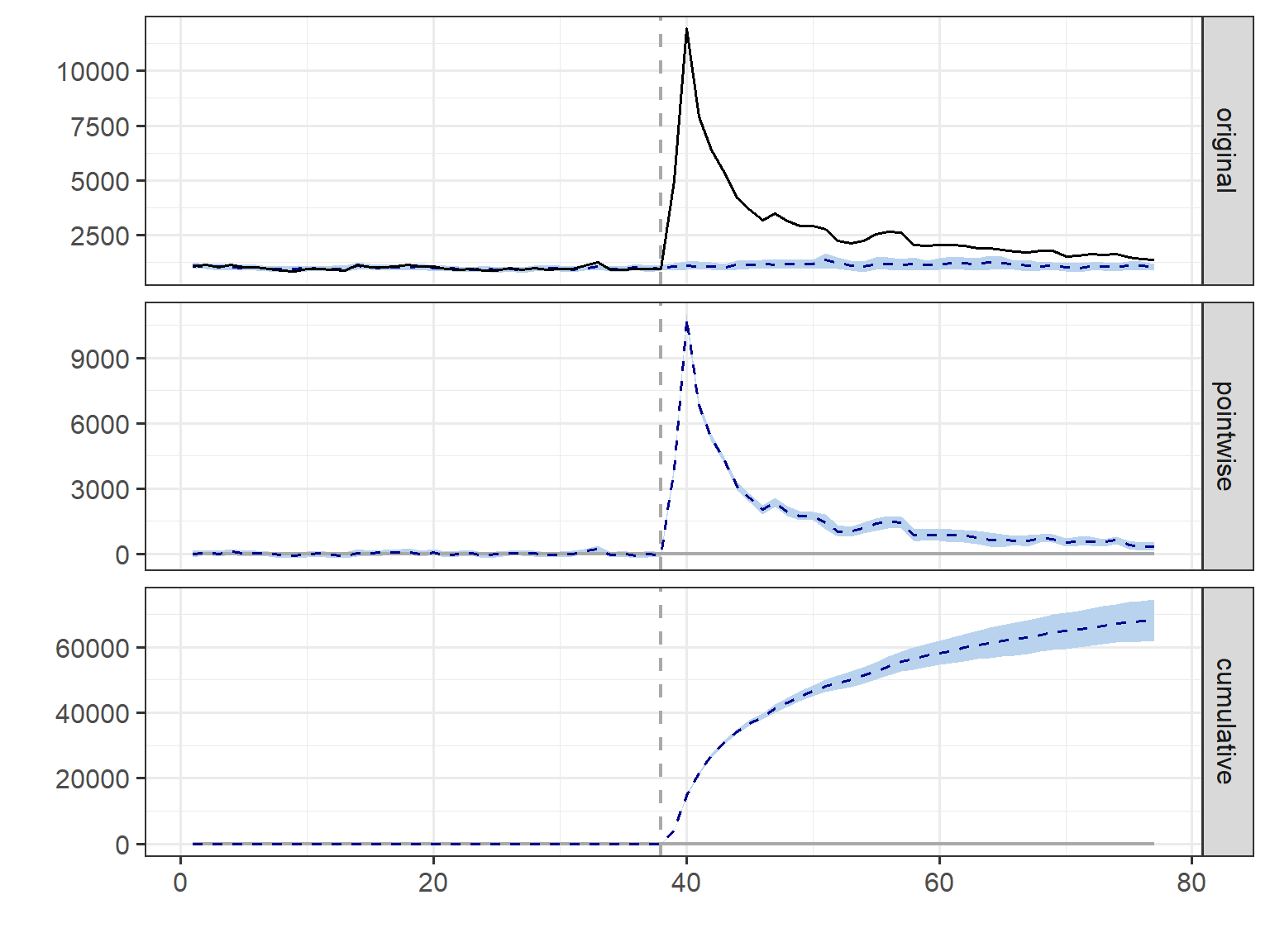
Summaries of the results
Code
summary(ci_m1)Posterior inference {CausalImpact}
Average Cumulative
Actual 2871 111962
Prediction (s.d.) 1122 (84) 43776 (3262)
95% CI [960, 1282] [37451, 50002]
Absolute effect (s.d.) 1748 (84) 68186 (3262)
95% CI [1589, 1911] [61960, 74511]
Relative effect (s.d.) 157% (20%) 157% (20%)
95% CI [124%, 199%] [124%, 199%]
Posterior tail-area probability p: 0.001
Posterior prob. of a causal effect: 99.8997%
For more details, type: summary(impact, "report")Code
summary(ci_m1, "report")Analysis report {CausalImpact}
During the post-intervention period, the response variable had an average value of approx. 2.87K. By contrast, in the absence of an intervention, we would have expected an average response of 1.12K. The 95% interval of this counterfactual prediction is [0.96K, 1.28K]. Subtracting this prediction from the observed response yields an estimate of the causal effect the intervention had on the response variable. This effect is 1.75K with a 95% interval of [1.59K, 1.91K]. For a discussion of the significance of this effect, see below.
Summing up the individual data points during the post-intervention period (which can only sometimes be meaningfully interpreted), the response variable had an overall value of 111.96K. By contrast, had the intervention not taken place, we would have expected a sum of 43.78K. The 95% interval of this prediction is [37.45K, 50.00K].
The above results are given in terms of absolute numbers. In relative terms, the response variable showed an increase of +157%. The 95% interval of this percentage is [+124%, +199%].
This means that the positive effect observed during the intervention period is statistically significant and unlikely to be due to random fluctuations. It should be noted, however, that the question of whether this increase also bears substantive significance can only be answered by comparing the absolute effect (1.75K) to the original goal of the underlying intervention.
The probability of obtaining this effect by chance is very small (Bayesian one-sided tail-area probability p = 0.001). This means the causal effect can be considered statistically significant. Code
plot(ci_m1$model$bsts.model, "coefficients")